预约演示
更新于:2025-12-05

Indian Council of Medical Research
更新于:2025-12-05
概览
标签
感染
肿瘤
其他疾病
预防性疫苗
mRNA
小分子化药
疾病领域得分
一眼洞穿机构专注的疾病领域
暂无数据
技术平台
公司药物应用最多的技术
暂无数据
靶点
公司最常开发的靶点
暂无数据
| 排名前五的药物类型 | 数量 |
|---|---|
| 预防性疫苗 | 5 |
| 小分子化药 | 4 |
| 化学药 | 2 |
| mRNA | 1 |
| 抗体偶联核素 | 1 |
关联
13
项与 Indian Council of Medical Research 相关的药物靶点 |
作用机制 EPRS抑制剂 |
在研适应症 |
非在研适应症- |
最高研发阶段临床前 |
首次获批国家/地区- |
首次获批日期- |
靶点- |
作用机制- |
在研适应症 |
非在研适应症- |
最高研发阶段临床前 |
首次获批国家/地区- |
首次获批日期- |
靶点- |
作用机制- |
在研适应症 |
非在研适应症- |
最高研发阶段临床前 |
首次获批国家/地区- |
首次获批日期- |
866
项与 Indian Council of Medical Research 相关的临床试验NCT07075315
National Collaborative Centre for Hepatic Regenerative Medicine (NC-CHRM): To Study the Safety and Efficacy of Mesenchymal Stem-Cell Therapy in the Management of Non-viral ACLF Patients: Phase-III Clinical Study
Liver disease deaths are rising, but transplants remain scarce in India. With over 100,000 needed annually and only
2,500 performed, non-transplant options are urgently needed. Regenerative therapy, especially mesenchymal stem cells (MSCs), shows promise but lacks validation, particularly for non-viral ACLF. The proposed NC-CHRM aims to develop and validate MSC-based therapy to promote native liver regeneration and offer a safe, effective, transplant-free treatment.
2,500 performed, non-transplant options are urgently needed. Regenerative therapy, especially mesenchymal stem cells (MSCs), shows promise but lacks validation, particularly for non-viral ACLF. The proposed NC-CHRM aims to develop and validate MSC-based therapy to promote native liver regeneration and offer a safe, effective, transplant-free treatment.
开始日期2028-01-01 |
申办/合作机构 |
NCT07131306
National Collaborative Centre for Hepatic Regenerative Medicine (NC-CHRM): To Study the Safety and Efficacy of Mesenchymal Stem-Cell (MSC) Therapy in the Management of Non-viral ACLF Patients: Phase-II Clinical Study
Liver disease deaths are rising, but transplants remain scarce in India. With over 100,000 needed annually and only
2,500 performed, non-transplant options are urgently needed. Regenerative therapy, especially MSCs, shows promise but lacks validation, particularly for non-viral ACLF. The proposed NC-CHRM aims to develop and validate MSC-based therapy to promote native liver regeneration and offer a safe, effective, transplant-free treatment.
2,500 performed, non-transplant options are urgently needed. Regenerative therapy, especially MSCs, shows promise but lacks validation, particularly for non-viral ACLF. The proposed NC-CHRM aims to develop and validate MSC-based therapy to promote native liver regeneration and offer a safe, effective, transplant-free treatment.
开始日期2027-01-01 |
申办/合作机构 |
CTRI/2025/07/090063
Assessment of cytogenic damage in chronic periodontitis and type 2 diabetes mellitus subjects through micronucleus test - NIL
开始日期2026-06-10 |
100 项与 Indian Council of Medical Research 相关的临床结果
登录后查看更多信息
0 项与 Indian Council of Medical Research 相关的专利(医药)
登录后查看更多信息
5,605
项与 Indian Council of Medical Research 相关的文献(医药)2026-02-01·BIOORGANIC & MEDICINAL CHEMISTRY LETTERS
Coumarin–benzimidazole hybrids: Design, synthesis and identification of potential anticancer agents
Article
作者: Kallingal, Anoop ; Sivapriya, M S ; Arya, C G ; Amrutha, P G ; Thomas, Neethu Mariam ; Banothu, Janardhan ; Gondru, Ramesh ; Chandrakanth, Munugala
A series of coumarin-benzimidazole conjugates (6a-p) was designed and synthesized via a polyphosphoric acid (PPA)-mediated condensation of o-phenylenediamine derivatives with coumarin-3-carboxylic acids. The in vitro anticancer activity of all conjugates was evaluated against lung (A549) and breast (MCF7) cancer cell lines. Except for hybrids 6l (A549) and 6c (MCF7), all compounds exhibited notable cytotoxicity toward both cell lines, with IC50 values ranging from 0.85 ± 0.07 to 48.01 ± 0.45 μM. Among them, the unsubstituted derivative 6b emerged as the most potent compound, showing IC₅₀ values of 0.85 ± 0.07 μM (A549) and 1.90 ± 0.08 μM (MCF7). Compounds 6a, 6c, and 6g also demonstrated strong activity against A549 cells, with IC50 values of 1.86 ± 0.19 μM, 1.05 ± 0.09 μM, and 2.23 ± 0.13 μM, respectively. Furthermore, the potent conjugates 6b and 6g were found to be non-toxic toward Vero cells (LD50 > 100 μM), indicating selective cytotoxicity toward cancer cells. In silico molecular docking studies suggested that the activity of compound 6b against both cell lines may result from inhibition of sphingosine kinase 1 (SK1), whereas the potency of 6g against A549 cells could be attributed to cyclin-dependent kinase 2 (CDK2) inhibition. These computational predictions warrant further validation through target-specific biological assays. ADMET analysis further revealed that conjugate 6b possesses favorable pharmacokinetic properties, supporting its potential as a promising lead for future anticancer drug development.
2026-01-01·CLINICA CHIMICA ACTA
Ethnicity-based variations in biological reference interval- A systematic scoping review
Article
作者: Chatterjee, Nabendu Sekhar ; Makhija, Ishika ; Bansal, Savita ; Maurya, Saanvi ; Chandra, Nilesh ; Sivakumar, Sasidharan ; Bhagat, Ruchika
BACKGROUND:
Biological reference intervals (RIs) are fundamental tools in clinical diagnostics, traditionally derived from geographically and ethnically homogeneous populations, predominantly of Western origin. Such generalized RIs often fail to account for variations arising from genetic, environmental, and lifestyle factors, which can impact clinical decision-making and contribute to health inequities, particularly in countries across Africa, India, and many other Southeast Asian nations. This scoping review investigates ethnicity-based variations in RIs for a range of biomarkers to highlight the importance of population-specific RIs.
METHODS:
Adhering to the Joanna Briggs Institute (JBI) guidelines, this scoping review examined the literature on ethnicity-based RI variations across multiple biomarkers, including Von Willebrand factor, C-reactive protein, thyroid-stimulating hormone, albumin, creatinine, and more. Studies were identified via searches in MEDLINE (via PubMed), Embase, Scopus, and Web of Science up to December 30, 2024. Eligibility was determined using the Population-Concept-Context (PCC) framework, focusing on observational studies analysing ethnicity-specific differences. Data extraction and charting were performed using CADIMA software, with independent review by two authors to ensure consistency.
RESULTS:
Out of 4,514 articles, a total of 15 studies met the inclusion criteria, encompassing multi-ethnic populations. Significant variations in biomarker levels were observed across ethnic groups, highlighting the inadequacy of generalized RIs. Notable differences included variations in lipid profiles, vitamin B12, and anti-Mullerian hormone levels. Regional distribution analysis highlighted gaps in research from underrepresented ethnic populations.
CONCLUSION:
This review emphasizes the critical need for ethnicity-specific RIs to improve diagnostic accuracy and promote equitable healthcare outcomes. Further research should focus on developing robust methodologies for establishing inclusive and representative RIs.
2026-01-01·Journal of Infection and Public Health
Malaria epidemiology, clinical characteristics, and concurrent infections in Delhi, India: A hospital-based study
Article
作者: Goel, Anandi ; Matlani, Monika ; Singh, Vineeta ; Haider, Mehvash ; Kojom Foko, Loick Pradel ; Hawadak, Joseph
OBJECTIVE:
Malaria remains a public health concern in India. Here, we analysed temporal trends, clinical features, concurrent infections, and determinants of malaria in patients in Delhi-based government hospital.
METHODS:
A longitudinal study was conducted between July 2022 and November 2023 in febrile patients attending a government hospital in Delhi. Clinical and biological examinations were done for each patient and screened for malaria parasites by rapid malaria antigen test and light microscopy, while concurrent infections were assessed by serological tests.
RESULTS:
A total of 4259 febrile patients were included among whom 87 (2.04 %) were infected with Plasmodium vivax and Plasmodium falciparum malaria species involved in malaria cases. The clinical presentation of cases was diverse, with a dominance of chills (80.46 %), hepatosplenomegaly (64.37), myalgia (56.32 %), generalised body ache (54.02 %), and jaundice/icterus (51.72 %). A few P. vivax- and P. falciparum-induced severe malaria cases were reported, with severe anaemia being the most frequent severe form. Nearly 45 % of malaria cases were co-infected with another pathogen (i.e., dengue virus, chikungunya virus, and Salmonella typhus). The climatic period July - September (aOR = 2.18; p < 0.05) and being male (aOR = 1.42; p < 0.05) were risk factors for malaria infection.
CONCLUSION:
Plasmodium vivax and Plasmodium falciparum were found in 2.07 % patients with a slight increase between 2022 and 2023. None of the factors tested were found to be associated with co-infections (dengue, chikungunya, and typhoid fever) or severe malaria, but in contrast, the climatic period July - September and being male were risk factors for malaria infection.
67
项与 Indian Council of Medical Research 相关的新闻(医药)2025-11-24
Mumbai:
Kiran Mazumdar-Shaw
, executive chairperson of biotech giant
Biocon
and the voice of the Indian biotechnology industry, has proposed sweeping reforms in the country’s drug regulatory functioning to boost research and innovation of new drugs and emerging treatment modalities.
A note from Shaw to
Niti Aayog
, India’s apex public policy think tank, calls for a dual-agency model that bifurcates the examination and approval process for newly researched drugs and devices between the
Indian Council of Medical Research
(
ICMR
), the premier medical research agency and the regulator Central Drugs Standard Control Organisation (
CDSCO
).
At present, Subject Expert Committees (SEC), a group of experts from diverse backgrounds ranging from key research and regulatory agencies and medical institutions approve new applications for clinical trials. Their recommendations are studied by another apex body before a final approval is granted by the Drug Controller General of India. But Shaw’s proposal seeks to empower
ICMR
to take up pre-licensing scientific reviews, which is a rigorous evaluation process to determine the safety and efficacy of new drugs.
Following the clearance based on scientific rationale, Shaw proposes the dossier (application) to move to
CDSCO
for final regulatory approval and licensing, including site inspections, GMP (good manufacturing practices) validation, and pharmacovigilance oversight.
“With its deep scientific, epidemiological, and translational research expertise,” Shaw’s note adds, “ICMR should be designated as the primary body for evaluating and approving clinical trial protocols for new drugs, devices, vaccines, and diagnostics.” She says ICMR’s domain scientists and ethics committees can ensure faster, more informed reviews - especially for investigator-initiated or early-stage trials, adding “This separation of responsibilities — scientific review (from ICMR) and regulatory licensing (from CDSCO) — would bring clarity, speed, and accountability to the system.”
“ICMR will frontend the scientific, ethical, and methodological evaluation of clinical trial protocols, issuing a Scientific Determination and Ethics Concurrence (SDEC) for early-phase studies such as First-in-Human (FIH) (clinical) trials for novel biologics, cell and gene therapies, RNA and precision-based therapeutics, new vaccines, complex diagnostics and investigator-initiated research,” Shaw’s proposal explains.
To keep the balance between the two powerful agencies, ICMR will co-opt nominated members from CDSCO to be members of the SDEC(s) as may be the case, while the joint body will operate with defined timelines and SOP-based dossier handling, with CDSCO representation to ensure alignment.
By
Vikas Dandekar
,

临床申请
2025-10-05
Dehradun: The Uttarakhand government on Saturday launched raids on medical stores and wholesale drug dealers across the state in the wake of
child deaths
in Madhya Pradesh and Rajasthan allegedly due to contaminated cough syrups.
Uttarakhand Health Secretary and FDA Commissioner Dr R Rajesh Kumar said
drug inspectors
in all districts have been ordered to collect samples of cough syrups from hospitals and shops in a phased manner within the month and have their quality laboratory tested so that any defective or harmful drugs can be immediately removed from the market.
Chief Medical Officers of the state have been ordered to implement the advisory received from the central government with immediate effect.
Central drug regulator Central Drugs Standard Control Organisation (
CDSCO
) has initiated risk-based inspections of drug manufacturing units in six states following collection of 19 samples including that of cough syrups, antipyretics and antibiotics.
The state health secretary said the safety and health of children as of paramount importance.
He urged all doctors in the state to take cognisance of the central government's advisory and not prescribe banned cough syrups for children.
"If doctors prescribe these syrups, medical stores will also sell them. Therefore, it is important that doctors themselves exercise responsibility and refrain from prescribing banned medicines," he said.
According to the central government's advisory, children under two years of age should not be given any type of cough or cold medicine without a doctor's advice, while the general use of these medicines is also not recommended for children under five years of age.
Kumar said the government has specifically banned syrups containing dextromethorphan and medicines containing a combination of chlorpheniramine maleate and phenylephrine hydrochloride for children under four years of age.
Following the order, raids were launched across the state on a war footing under the leadership of FDA Additional Commissioner and Drug Controller Tajbar Singh Jaggi. The additional commissioner personally inspected drug stores in several areas of Dehradun, including Jogiwala and Mohkampur.
He said that if any defects are found at any level, strict legal action will be taken against the concerned company or seller.
Jaggi also appealed to the public to consult a doctor before giving any medicine to children, and to immediately contact the nearest health centre or hospital if any adverse effects are observed after taking any medicine.
The inspections by the CDSCO began on Friday with the aim to identify gaps that may have led to drug quality failures and also suggest process improvement to avoid such incidents in future, the ministry said.
Additionally, a multidisciplinary team comprising experts from the National Institute of Virology, the Indian Council of Medical Research, National Environmental Engineering Research Institute, CDSCO and AIIMS-Nagpur, among others, are still analysing the various samples and factors to assess the cause of deaths in and around Chhindwara in Madhya Pradesh, it stated.
The manufacturing units are in Himachal Pradesh, Uttarakhand, Gujarat, Tamil Nadu, Madhya Pradesh and Maharashtra where 19 samples of different drugs were manufactured, sources in the Union health ministry said. PTI
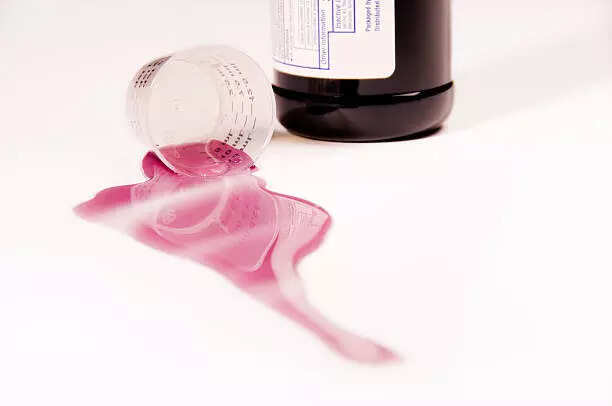
2025-10-05
New Delhi: The Central drug regulator CDSCO has initiated risk-based inspection of drug manufacturing units in six states following collection of 19 samples including that of
cough
syrups, antipyretics and antibiotics in the wake of child deaths in
Madhya Pradesh
and Rajasthan allegedly due to contaminated cough syrups.
The manufacturing units are in Himachal Pradesh, Uttarakhand, Gujarat,
Tamil Nadu
, Madhya Pradesh and Maharashtra where 19 samples of different drugs were manufactured, sources in the health ministry said.
The inspections by the Central Drugs Standard Control Organisation (CDSCO) began on Friday with the aim to identify gaps that may have led to drug quality failures and also suggest process improvement to avoid such incidents in future, the ministry said.
Additionally, a multidisciplinary team comprising experts from the National Institute of Virology, the Indian Council of Medical Research, National Environmental Engineering Research Institute, CDSCO and AIIMS-Nagpur, among others, are still analysing the various samples and factors to assess the cause of deaths in and around Chhindwara in Madhya Pradesh, it stated.
On Friday, the ministry said that six samples that have tested by CDSCO and three by the Madhya Pradesh Food and Drugs Administration (MPFDA) were found to be free of Diethylene Glycol (DEG) and Ethylene Glycol (EG) contaminants that are known to cause serious kidney injury.
Sources from the ministry clarified that the "samples that have been tested so far by CDSCO were not of the two suspected cough syrups one of which is
Coldrif
that have been under scanner since the deaths".
"The six drug samples tested by CDSCO which did not show presence of DEG/EG were of other drugs and syrups including antibiotics, antipyretics and Ondansetron, consumed by the children who fell ill in the Chhindwara district of Madhya Pradesh," a source said.
The analysis of the samples of Coldrif and the other suspected cough syrup by Madhya Pradesh state drug authorities is still underway.
At the request of the Madhya Pradesh government, the Tamil Nadu Food Safety and Drug Administration Department tested samples of Coldrif Cough Syrup collected from the manufacturing unit of
Sresan Pharma
in Kanchipuram, Tamil Nadu.
"The results were shared with us late Friday evening... The samples had DEG beyond the permissible limit," the ministry said.
Following this, the Tamil Nadu government banned the sale of the cough syrup and ordered its removal from the market.
Inspections were also conducted at the pharmaceutical company's manufacturing facility in Sunguvarchathram in neighbouring Kancheepuram district during the last two days, and samples have been collected, he said.
Madhya Pradesh government has also written to Himachal Pradesh Drug Authority for conducting testing of the other cough syrup under scanner.
The Madhya Pradesh government also on Saturday banned the sale of Coldrif syrup.
According to officials, nine children have died due to suspected renal failure since September 7. Currently, 13 children, including eight from Chhindwara and Nagpur, are undergoing treatment.
"The deaths of children in Chhindwara due to Coldrif syrup are extremely tragic. The sale of this syrup has been banned across Madhya Pradesh. A ban is also being imposed on other products of the company that manufactures the syrup," Chief Minister
Mohan Yadav
stated on X.
The syrup was manufactured at a factory in Kanchipuram. Following the incident, the state government requested that the Tamil Nadu government conduct an inquiry. The investigation report was received this morning, and strict action has been taken, he stated.
A team has also been constituted at the state level to investigate the matter, Yadav said, adding that the guilty will not be spared.
Meanwhile, Kerala Drugs Control department also has suspended the sale of Coldrif cough syrup in the state following reports from other states that flagged issues with one of the batches of the Coldrif syrup, Kerala Health Minister
Veena George
on Saturday said.
A preliminary inquiry by the State Drugs Control Department revealed that the flagged batch of the drug was not sold in Kerala, the minister clarified in a statement.
Noting that the Drugs Control department has conducted intensive inspections in the state, George said samples of Coldrif syrup have been collected for examination.Along with this, samples of other cough syrups are also being examined.
Besides, test samples of cough syrups collected from a Chennai-based firm during an inspection conducted at its facility have been found to be 'adulterated', an official of the Food Safety and Drug Administration department said on Saturday.
The officials have sought an explanation from the Tamil Nadu-based company and has instructed it to halt production at its facility near here, the official said.
The development came after Tamil Nadu government banned the sales of cough syrup 'Coldrif' and ordered removal of stocks of the medicine from the market.
Apart from that, the Rajasthan government has suspended the state drug controller and halted the distribution of medicines manufactured by Jaipur-based company
Kaysons Pharma
, officials said.
The Medical and Health Department stopped the supply of all 19 medicines manufactured by Kaysons Pharma until further orders, they said.
Distribution of all other cough syrups containing Dextromethorphan has also been suspended.
The government has placed drug controller Rajaram Sharma under suspension for allegedly influencing the process of determining drug standards, the department said.
This comes amid reports of deaths of 11 children, nine in Madhya Pradesh and two in Rajasthan, allegedly linked to contaminated cough syrup.
Chief Minister Bhajanlal Sharma has ordered a detailed probe into the matter along with effective action.
Following his instructions, an expert committee is being constituted to investigate the issue.
The state has reiterated the advisory, while the Drug Controller General of India on Friday advised that the syrup should normally be given only to children above five years, and in no circumstances to those under two years of age.
Officials added that drugs which are potentially harmful to children and pregnant women will now carry clear warning labels.
In Uttarakhand, joint teams of the health department and Food Safety and Drug Administration (FDA) are conducting raids on medical stores, wholesale drug vendors and hospital pharmacies in all districts.
According to the managing director of Rajasthan Medical Services Corporation Ltd (RMSCL), Pukhraj Sen, over 10,000 samples of Kaysons Pharma drugs have been tested since 2012, of which 42 failed the quality standards.
As a precaution, the supply of all 19 medicines manufactured by the company has been suspended.
The Centre on Friday had issued an advisory to all states saying that cough and cold medications should not be prescribed to children below two years.
The DGHS, Union Health ministry in its advisory mentioned that Cough syrups are generally not recommended for ages below 5 years and above that, any use should follow careful clinical evaluation with close supervision and strict adherence to appropriate dosing, the shortest effective duration and avoiding multiple drugs combinations. PTI

100 项与 Indian Council of Medical Research 相关的药物交易
登录后查看更多信息
100 项与 Indian Council of Medical Research 相关的转化医学
登录后查看更多信息
组织架构
使用我们的机构树数据加速您的研究。
登录
或

管线布局
2026年03月01日管线快照
管线布局中药物为当前组织机构及其子机构作为药物机构进行统计,早期临床1期并入临床1期,临床1/2期并入临床2期,临床2/3期并入临床3期
药物发现
3
10
临床前
其他
3
登录后查看更多信息
当前项目
登录后查看更多信息
药物交易
使用我们的药物交易数据加速您的研究。
登录
或

转化医学
使用我们的转化医学数据加速您的研究。
登录
或

营收
使用 Synapse 探索超过 36 万个组织的财务状况。
登录
或

科研基金(NIH)
访问超过 200 万项资助和基金信息,以提升您的研究之旅。
登录
或

投资
深入了解从初创企业到成熟企业的最新公司投资动态。
登录
或

融资
发掘融资趋势以验证和推进您的投资机会。
登录
或

生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用

