预约演示
更新于:2025-12-18

The New Zealand Institute for Plant & Food Research Ltd.
更新于:2025-12-18
概览
关联
51
项与 The New Zealand Institute for Plant & Food Research Ltd. 相关的临床试验ACTRN12624000587505
Pilot Study Investigating Relationship between Habitual Diet and Response to Acute Stress or Relaxation in Healthy Adults
开始日期2024-05-02 |
ACTRN12623000045617
The immune response to consuming probiotic strains in healthy adults: a randomised double-blind, placebo-controlled, parallel clinical study
开始日期2024-03-01 |
ACTRN12621000774820
Investigating the effect of BerriQi® Boysenberry and apple product consumption on lung function and immune protection following acute ozone exposure induced lung inflammation in healthy volunteers.
开始日期2023-01-02 |
100 项与 The New Zealand Institute for Plant & Food Research Ltd. 相关的临床结果
登录后查看更多信息
0 项与 The New Zealand Institute for Plant & Food Research Ltd. 相关的专利(医药)
登录后查看更多信息
2,513
项与 The New Zealand Institute for Plant & Food Research Ltd. 相关的文献(医药)2026-01-01·Methods in molecular biology (Clifton, N.J.)
Pilosella: A Dicotyledonous Model for Studying Aposporous, Autonomous Apomixis
Article
作者: Koltunow, Anna M G ; Bicknell, Ross
Pilosella, a member of the Compositae, is a model system used to study the molecular genetics of aposporous apomixis. These plants are small, rapidly growing perennials that are easy to cultivate both in the greenhouse and in tissue culture. Apomixis in Pilosella occurs by apospory where mitotically derived embryo sacs arise adjacent to cells undergoing female gamete meiosis in the ovule. Seed initiation is autonomous, where both embryo and endosperm form without fertilization in the aposporous embryo sac. Apomixis is not fully penetrant in Pilosella. Instead, plants are facultatively apomictic, and apomixis can be easily scored through the simple decapitation of the immature capitulum bud. Natural sexual and facultatively apomictic forms are readily cross-compatible, facilitating comparative studies of inheritance and allele function. A wide range of experimental methods have been described for these plants, including histological techniques for studying the cytological aspects of apomixis, an efficient Agrobacterium-mediated transformation system, CRISPR/Cas9 mutagenesis, and mapping approaches that use deletion mutation and segregation in polyhaploid populations. Freely available online resources include a genome assembly, a molecular map based on cDNA markers and a transcriptome database. Collectively, these resources make Pilosella a highly tractable experimental system for studying the genetic control of native apomixis.
2026-01-01·Methods in molecular biology (Clifton, N.J.)
Analysis of Gametophytic Apomixis Using Confocal Microscopy
Article
作者: Banfi, Camilla ; Barrell, Philippa Jane
Apomixis is an asexual reproductive mechanism that takes place deeply inside the female reproductive organs of the plant, in ovules and seeds. In gametophytic apomixis, an unreduced female gametophyte is produced by a modified meiosis of the megaspore mother cell (dipolspory) or from a somatic initial cell (apospory). The unreduced, nonrecombined egg cell develops subsequently into an embryo by parthenogenesis. The cyto-embryological study of apomixis is challenging because of the inaccessibility of these structures. Consequently, images of apomeiosis and parthenogenesis with high definition are limited to a few species. In this chapter, we show the application of a Feulgen staining protocol combined with confocal microscopy for the study of nonreductional megasporogenesis and autonomous embryo formation in diplosporous apomictic Taraxacum officinale and aposporous apomictic Pilosella piloselloides var. praealta. Using a rapid and technically simple method, performed on whole-mount ovaries, we have obtained high-resolution images of the female reproductive cells. Furthermore, we highlight the application of this protocol for the study of loss-of-diplospory and loss-of-parthenogenesis mutants in the same species.
2026-01-01·Methods in molecular biology (Clifton, N.J.)
Genetic Mapping in Apomicts
Article
作者: Catanach, Andrew ; Winefield, Chris ; Bicknell, Ross
Genetic mapping efforts in apomicts date back several decades, but significant progress only became possible with the application of molecular mapping tools. In this chapter, we present the approaches that have been taken to apply these techniques to apomictic species, the necessary experimental steps, caveats associated with interpreting the data, and potential new marker approaches. A brief overview of our understanding of the genetics of natural apomixis is also presented.
3
项与 The New Zealand Institute for Plant & Food Research Ltd. 相关的新闻(医药)2025-05-28
ALAMEDA, Calif. , May 28, 2025 (GLOBE NEWSWIRE) -- genXtraits Inc. (genXtraits) today announced that it has expanded its license and collaboration agreement with The New Zealand Institute for Plant and Food Research Limited (Plant & Food Research) to apply proprietary technology for increasing Vitamin C levels in crops. genXtraits and Plant & Food Research entered an agreement in late 2023 under which genXtraits received an option to obtain an exclusive worldwide license under Plant & Food Research’s patents to develop and commercialize certain crops with the elevated vitamin C trait. Having met contractual requirements over the past 18 months, genXtraits has now exercised its exclusive commercial license, as well as broadening the scope of licensed applications. genXtraits’ expanded license includes major row crops, such as corn and soybean, as well as a broad range of bioenergy crops in addition to the original suite of licensed species, which included tomato, potato, peppers and eggplant. Plant & Food Research has retained commercial rights in specific areas, for example including applications in pipfruit, kiwifruit, berryfruit and grapes. The base license comprises genetic trait technology for elevating the levels of vitamin C in plant cells, which was co-developed by Dr. Roger Hellens, the co-founder of genXtraits, when he was previously a project leader at Plant & Food Research. “Vitamin C is a very potent cellular protectant against oxidative damage,” said Dr. Hellens. “This particular trait provides a double whammy in terms of its benefits. Higher levels of vitamin C in foods like tomatoes and potatoes can provide a dietary benefit, since the compound promotes iron uptake by the human gut. This could help combat anemia, which is a major problem in many countries, especially those with a predominantly vegetarian diet. Additionally, Vitamin C defends crop cells against stress caused by drought and disease, which means it can also act as a yield stability trait that protects overall crop yield.” Dr. Peter Cook, General Manager Business Development at Plant & Food Research noted, “We are delighted to expand our partnership with genXtraits. genXtraits has established a unique technical platform, which can deliver novel traits through gene edits that increase the levels of regulatory proteins in crop cells. Their platform has already delivered improvements to the base inventions in the patents owned by Plant & Food Research. Under the terms of the deal, Plant & Food Research enjoys a license-back under this new IP from genXtraits for application in its core crops.” Dr. Oliver Ratcliffe, CEO of genXtraits commented, “Given the complementarity of genXtrait’s platform with the technology licensed from Plant & Food Research, it made perfect sense to broaden our collaboration. Our shared goal is to implement this important trait in as many crops as possible. To this end, we are looking to add additional partners to participate in our collaboration, especially for applications in corn, soybean, and potatoes and we are actively seeking licensees for the vitamin C trait.” About genXtraits: genXtraits is a California-based crop genomics company, which was founded in late 2022 by plant biotechnology industry veterans. The firm’s unique selling point is its STRM™ platform for the delivery of dominant genetic traits, which stem from simple gene edits that elevate the cellular levels of important regulatory proteins. The company is focused on developing the next generation of crop traits, which will both protect crop yield in the face of climate change and provide consumers with more delicious and healthy food choices. For more information, please visit: https://genxtraits.com/ Contact: Dr. Oliver J. Ratcliffe (contact@genxtraits.com) About Plant & Food Research: The New Zealand Institute for Plant and Food Research Limited is a Crown Research Institute established under the Crown Research Institutes Act 1992. Plant & Food Research uses world-leading science to support the food sector in improving ways to grow, fish, harvest, prepare and share food. The organization has close to 1,000 people that work across New Zealand and the world to help deliver healthy foods from the world’s most sustainable systems. For more information visit: https://www.plantandfood.com Contact: Emma Timewell, Communications Manager (emma.timewell@plantandfood.co.nz)
引进/卖出
2023-03-02
·生物谷
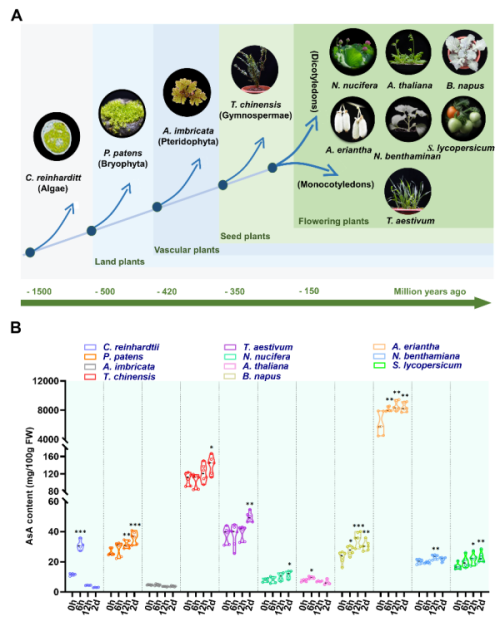
冷胁迫限制植物的生长、发育和分布,是影响农业生产持续稳定发展的重要因素之一。维生素C(L-抗坏血酸,AsA)是一种抗氧化剂,参与非生物应激耐受和活性氧(ROS)代谢。
冷胁迫限制植物的生长、发育和分布,是影响农业生产持续稳定发展的重要因素之一。维生素C(L-抗坏血酸,AsA)是一种抗氧化剂,参与非生物应激耐受和活性氧(ROS)代谢。探究低温胁迫如何诱导维生素C生物合成以减少对植物的氧化损伤,这对增强植物抗寒性、提高作物维生素C含量具有重要意义。
近日,中国科学院武汉植物园钟彩虹研究团队在Plant Physiology上,在线发表了题为Kiwifruit bZIP transcription factor AcePosF21 elicits ascorbic acid biosynthesis during cold stress的研究论文。该研究发现了植物在冷胁迫下诱导维生素C生物合成以减轻寒冷对植物氧化损伤现象,阐明了猕猴桃通过AceMYB102-AcePosF21-AceGGP3分子网络调控维生素C合成以减缓冷损伤的分子机制。
研究通过分析从单细胞到被子植物在冷胁迫下叶、果等组织的维生素C含量变化发现,冷胁迫短时间内能够诱导大部分植物维生素C的合成。利用高维生素C的猕猴桃为材料,研究发掘了一个bZIP家族的转录因子AcePosF21。该转录因子能够被低温信号触发,对猕猴桃维生素C的生物合成和防御反应具有重要的调控作用。此外,科研团队利用VIGS诱导的基因沉默或CRISPR/Cas9介导的编辑,证实了AcePosF21的缺失会降低猕猴桃维生素C浓度,增加ROS的生成。研究进一步发现:AcePosF21能够与R2R3-MYB转录因子AceMYB102相互作用,直接结合到维生素C合成的关键基因AceGGP3启动子区、激活AceGGP3基因的表达;AceGGP3基因的高表达能够促进维生素C的合成并清除冷胁迫诱导的过量ROS,最终减缓猕猴桃冷损伤。
本研究证实,维生素C作为重要的抗氧化物质,不仅为人类提供维持健康所需的重要营养物质,而且在植物中能够响应胁迫、参与抗寒等生理过程,为猕猴桃的低温抗性育种提供了基因资源和重要理论支撑。
研究工作得到中科院“种子精准设计与创造”先导专项、湖北省洪山实验室、国家重点研发计划、中科院青年创新促进会和国家作物表型组学研究(神农)基地的支持。新西兰植物与食品研究所的科研人员参与研究。
图1.不同进化地位的植物(A)在冷胁迫下AsA浓度(B)的变化
临床研究
2022-08-11
A research team has developed a sensing technology that can detect whether a substance was exposed to light using metahologram.
During the COVID-19 pandemic last year, an incident occurred where vaccines exposed to room temperature had to be discarded. Biomedical substances, including vaccines, risk deterioration if not stored properly, so strict management is required during production and storage. In particular, exposure to light may reduce the vaccine's efficacy, so it is important to check whether it has been damaged by light exposure.
Recently, a POSTECH research team led by Professor Junsuk Rho (Department of Mechanical Engineering and Department of Chemical Engineering) and Ph.D. candidate Joohoon Kim (Department of Mechanical Engineering) has collaborated with Dr. Jonghyun Choi (The New Zealand Institute for Plant and Food Research Ltd.) to propose a novel sensor platform that detects light exposure using a "universal metahologram," which can not only operate under all states of polarizations, but also transmit multiple lights.
A metahologram made of metamaterials is a technology that displays different holographic images depending on the direction of incident light. Its applicability in next-generation displays and as security sensor was highly anticipated as it can display multiple images simultaneously.
However, a limitation existed in which a metahologram that operates in all incident polarizations could only express a limited number of colors. Conversely, metaholograms that show different colors only respond to certain incident polarizations.
To overcome this limitation, the research team successfully fabricated a metahologram that emits brilliant colors regardless of any incident light. Moreover, it was confirmed that this new metahologram can be used as a sensor when attached to a container designed for storing a chemical or biomaterial to detect light exposure. The research team particularly increased the possibility of large commercial manufacturing for metaholograms by developing a printing process that replicates them over flexible substrates.
The research findings can be useful for secure packaging and transportation of light-sensitive chemical or biomedical substances. The technology can be applicable in intelligent packaging and labeling to prevent counterfeits and to verify the authenticity of agricultural and fishery products such as honey and kiwi, or secondary processed foods exported by New Zealand.
Recently published in ACS Applied Materials and Interfaces, this study was conducted with the support from the POSCO-POSTECH-RIST Convergence Research Center program funded by POSCO, LGD-SNU incubation program funded by LG Display, and from the South Korea-New Zealand Joint Research Partnership Program and the Strategic Nanomaterial Technology Development Program of the National Research Foundation of Korea.
疫苗合作
100 项与 The New Zealand Institute for Plant & Food Research Ltd. 相关的药物交易
登录后查看更多信息
100 项与 The New Zealand Institute for Plant & Food Research Ltd. 相关的转化医学
登录后查看更多信息
组织架构
使用我们的机构树数据加速您的研究。
登录
或

管线布局
2026年02月28日管线快照
无数据报导
登录后保持更新
药物交易
使用我们的药物交易数据加速您的研究。
登录
或

转化医学
使用我们的转化医学数据加速您的研究。
登录
或

营收
使用 Synapse 探索超过 36 万个组织的财务状况。
登录
或

科研基金(NIH)
访问超过 200 万项资助和基金信息,以提升您的研究之旅。
登录
或

投资
深入了解从初创企业到成熟企业的最新公司投资动态。
登录
或

融资
发掘融资趋势以验证和推进您的投资机会。
登录
或

生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用