预约演示
更新于:2025-05-07
Huntington Disease-Like Syndrome
亨廷顿病样综合征
更新于:2025-05-07
基本信息
别名 Huntington Disease-Like Syndrome、Huntington Disease-Like Syndromes、Huntington disease phenocopy syndrome + [7] |
简介- |
关联
2
项与 亨廷顿病样综合征 相关的药物靶点 |
作用机制 GPR88 激动剂 |
非在研适应症- |
最高研发阶段临床前 |
首次获批国家/地区- |
首次获批日期1800-01-20 |
靶点- |
作用机制 PET imaging |
在研适应症 |
非在研适应症- |
最高研发阶段临床前 |
首次获批国家/地区- |
首次获批日期1800-01-20 |
100 项与 亨廷顿病样综合征 相关的临床结果
登录后查看更多信息
100 项与 亨廷顿病样综合征 相关的转化医学
登录后查看更多信息
0 项与 亨廷顿病样综合征 相关的专利(医药)
登录后查看更多信息
119
项与 亨廷顿病样综合征 相关的文献(医药)2025-07-01·Archives of Medical Research
West-Central African Ancestry of the Repeat-Expansion Founder Mutation on the JPH3 Gene in Mexican Patients With Huntington's Disease-Like 2
Article
作者: Romero-Hidalgo, Sandra ; Monroy-González, Julio César Misael ; Macias-Kauffer, Luis Rodrigo ; Yescas-Gómez, Petra ; Díaz-Barba, Karina ; Ramírez-García, Miguel Ángel ; Castañeda-López, Gabriela ; Villarreal-Molina, María Teresa
2025-05-01·Journal of Neurology, Neurosurgery & Psychiatry
Huntington’s disease phenocopy syndromes revisited: a clinical comparison and next-generation sequencing exploration
Article
作者: Hensman Moss, Davina J ; Tabrizi, Sarah J ; Eberle, Michael ; Wild, Edward J ; Guntoro, Fernando ; Rosser, Anne Elizabeth ; Koriath, Carolin Anna Maria ; Flower, Michael ; Dolzhenko, Egor ; Mead, Simon ; Norsworthy, Penelope ; Hummerich, Holger
2025-02-01·Journal of Huntington's Disease
Comparative analysis of neurofilament light chain in Huntington's disease like 2 and Huntington's disease
Article
作者: Ferreira-Correia, Aline ; Wild, Edward J ; Krause, Amanda ; Byrne, Lauren M ; Anderson, David G ; Rodrigues, Filipe B
12
项与 亨廷顿病样综合征 相关的新闻(医药)2023-11-24
·药时代
2023年11月21日,默沙东(“MSD”)与Caraway Therapeutics(简称“Caraway”)共同宣布,两家公司已达成最终协议。根据协议条款:默沙东将通过子公司收购Caraway,总金额约6.1亿美元,包括未披露的预付款以及或有里程碑付款。
Caraway利用遗传数据和独特的生物学理解,来开发专有的小分子,激活细胞清除和回收过程-加速清除累积的有毒物质和有缺陷的细胞成分。
遗传学的新见解强调了溶酶体功能与多器官系统病理之间的联系,包括中枢神经系统(CNS)、心脏、肾脏和肌肉。以这些机制为靶点,Caraway为患有神经退行性疾病和罕见疾病(包括帕金森病(PD)、肌萎缩性侧索硬化症(ALS)和溶酶体沉积病(lsd)的患者提供潜在的疾病修饰化合物。
公司正在推进一系列强大的新型小分子疗法,用于治疗神经退行性疾病和罕见疾病,目前均处于临床前的阶段。
TRPML1:TRPML1是通道粘磷脂亚家族的成员,像其他TRP通道一样,TRPML1 具有六个跨膜螺旋 (S1~S6),以及一个由 S5、S6和 2 个孔螺旋 (PH1和PH2) 组成的孔区域。它是一种释放 Ca2+的阳离子通道,可以介导溶酶体的钙信号传导和稳态,也是转运和自噬相关事件的重要调节因子。
TRPML1的突变能够导致溶酶体贮积病黏液脂病 IV (MLIV) 的发生,会造成认知、语言和运动缺陷和视网膜变性等各种症状的发生,因此TRPML1作为治疗溶酶体贮积症的靶点,在临床上表现出巨大的潜能。
由于神经元对溶酶体贮积更为敏感,而且神经元是终末分化的细胞,一旦死亡后无法得到有效的补充,因此大部分溶酶体贮积症都出现神经系统退行性病变的症状,这一现象也将溶酶体与神经退行性疾病紧密联系起来。其可以分为急性神经退行性病和慢性神经退行性病,前者主要有中风、脑损伤;后者主要有亨廷顿病(HD)、帕金森病(PD)、阿尔兹海默病(AD)等。
TMEM175:TMEM175是一种新型的氢离子激活的氢离子通道(LyPAP),介导氢离子快速流出。TMEM175缺乏可导致溶酶体过度酸化,破坏蛋白水解活性,促进体内帕金森致病相关的α-突触核蛋白聚集。2022年6月,Cell杂志在线发表了美国密西根大学徐浩新(Haoxing Xu)团队的相关研究,证实在溶酶体生理条件下,TMEM175作为氢离子激活的氢离子通道介导溶酶体氢离子外流,平衡V-ATP酶介导的氢离子内流,以维持稳定的溶酶体pH值。此外,作为维持溶酶体最佳pH值的氢离子激活的氢离子通道TMEM175在细胞水平上调节溶酶体蛋白水解酶Cathepsin B和Cathepsin D活性,以及体内α-突触核蛋白的清除,为PD、AD等神经退行性疾病和LSDs的病理过程和治疗方法研究提供新思路。
2018年11月,Caraway前身Rheostat Therapeutics宣布完成2300万美元A轮融资,该轮融资由MRLV和AbbVie Ventures领投,包括Amgen Ventures,
Alexandria Venture Investments和Mayo Clinic。现有投资者SV Health investors和Dementia Discovery Fund跟投。
2021年6月,该公司曾与艾伯维达成一项合作,开发和商业化靶向TMEM175的小分子疗法,首付款1700万美元。
参考资料
1、公司官网
2、公众号:晶蛋生物、医药魔方
3、Hu M, Li P, Wang C, et al. Parkinson’s disease-risk protein TMEM175 is a proton-activated proton channel in lysosomes[J]. Cell, 2022, 185(13): 2292-2308. e20.
内容来源于网络,如有侵权,请联系删除。
并购
2023-11-06
·药明康德
Neurocrine Biosciences日前公布KINECT-HD2临床3期研究的中期结果,该研究评估Ingrezza(valbenazine)胶囊作为亨廷顿病(HD)相关舞蹈病成人患者长期治疗选择的作用。初步数据表明,早在第2周时每日一次Ingrezza胶囊给药即改善患者的舞蹈病症状,该获益持续至第50周。
KINECT-HD2的中期分析显示,在初始剂量40 mg下,Ingrezza在第2周起表现出疗效,并在高达80 mg的剂量下维持其疗效至第50周(参见下图)。
相当大比例的受试者(60.9%)及其临床医生(58.9%)报告,到第6周时症状状态“改善”或“极大改善”。到第50周,这些数值进一步增加,约四分之三的受试者(74.2%)和研究人员(76.9%)报告症状显著改善。
本试验中显示的安全性特征与既往结果一致。最常见治疗相关不良事件包括跌倒(30.4%)、疲乏(24.0%)和嗜睡(24.0%)。
Ingrezza是一款选择性囊泡单胺转运体2(VMAT2)抑制剂,对VMAT1、多巴胺能、5-羟色胺能、肾上腺素能、组胺能或毒蕈碱受体无明显亲和力。Ingrezza通过选择性靶向VMAT2,被认为可减少额外的多巴胺信号,这可能导致不可控运动的减少。Ingrezza已获FDA批准用于治疗成人亨廷顿病患者的迟发性运动障碍,并可与大多数精神科药物(如抗精神病药或抗抑郁药)联合使用。今年8月,Ingrezza获FDA批准用于治疗亨廷顿病相关舞蹈病成人患者。
参考资料:[1] Neurocrine Biosciences® Presents INGREZZA® (valbenazine) Capsules Interim Data Demonstrating Sustained Improvements in Chorea Associated With Huntington's Disease Through Week 50 at Huntington Study Group 2023.
Retrieved November 3, 2023 from https://neurocrine.gcs-web.com/news-releases/news-release-details/neurocrine-biosciencesr-presents-ingrezzar-valbenazine-capsules
内容来源于网络,如有侵权,请联系删除。
基因疗法临床结果
2023-10-26
·药明康德
今日,Alnylam Pharmaceuticals公布了在研RNAi疗法ALN-APP治疗阿尔茨海默病(AD)和脑淀粉样血管病(CAA)的临床1期试验的积极中期结果。试验结果显示,单剂ALN-APP的持续药代动力学活性可以长达10个月,同时观察到与阿尔茨海默病和脑淀粉样血管病相关的淀粉样蛋白片段Aβ42和Aβ40的显著降低。
Alnylam开发的C16偶联技术平台专门优化对中枢神经系统组织的递送。通过在RNAi上偶联不同的脂质分子,调整它们的亲脂性,这一系统在大鼠和非人灵长类模型中,已经能在广泛中枢神经系统(CNS)组织中显著降低mRNA表达。
ALN-APP是首款应用C16偶联技术,靶向编码APP蛋白的mRNA的临床期RNAi疗法,由Alnylam与再生元(Regeneron)共同合作开发。它可以通过降解mRNA,降低APP蛋白的表达。APP基因的突变是导致早发性阿尔茨海默病和脑淀粉样血管病的重要原因,而且淀粉样蛋白沉积是阿尔茨海默病患者大脑的标志性特征之一。
今日公布的试验结果显示,接受75 mg ALN-APP单剂治疗的患者脑脊液(CSF)中可溶性APPα(sAPPα)和可溶性APPβ(sAPPβ)快速和持续减少,最大降幅分别为84%和90%。这些效果具有高度持久性,在接受75 mg的ALN-APP单剂治疗后10个月,sAPPα和sAPPβ的水平仍然平均降低33%和39%。
此外,探索性分析显示,患者CSF中的Aβ42和Aβ40水平显著降低,这些可溶性淀粉样蛋白片段聚集在一起会形成阿尔茨海默病和脑淀粉样血管病患者体内出现的淀粉样蛋白沉积。在接受剂量为75 mg的ALN-APP单剂治疗2个月后,CSF中Aβ42和Aβ40水平平均降低49%和71%。
Alnylam表示将继续进行ALN-APP单剂量探索研究,试验的多剂量给药部分也已经启动,首位患者已经完成给药。
在2021药明康德全球论坛上,Alnylam公司时任首席运营官Yvonne Greenstreet女士曾强调构建创新治疗技术平台的重要性。她表示,优化创新技术平台需要很长的时间,通常,一项科学发现从最初获得诺贝尔奖和发表论文,到转化研发出首个药物并应用于临床,可能已经过去了20年。然而,一旦创新技术平台获得成功,后续新药开发的成功率就会变得特别高。Alnylam公司的RNAi平台经过多年发展,在2018年推出首款获批药物,而不久后的现在,业内已经有多款RNAi疗法获批。目前Alnylam公司的研发管线中,还有多款靶向CNS疾病靶点的在研疗法,治疗渐冻症(ALS),亨廷顿病,帕金森病等神经退行性疾病。期待RNAi疗法在治疗CNS疾病方面早日获得突破,造福广大患者。
参考资料:[1] Alnylam Reports Additional Positive Interim Phase 1 Results for ALN-APP, in Development for Alzheimer’s Disease and Cerebral Amyloid Angiopathy. Retrieved October 25, 2023, from https://www.businesswire.com/news/home/20231025380274/en
内容来源于网络,如有侵权,请联系删除。
临床结果信使RNA
分析
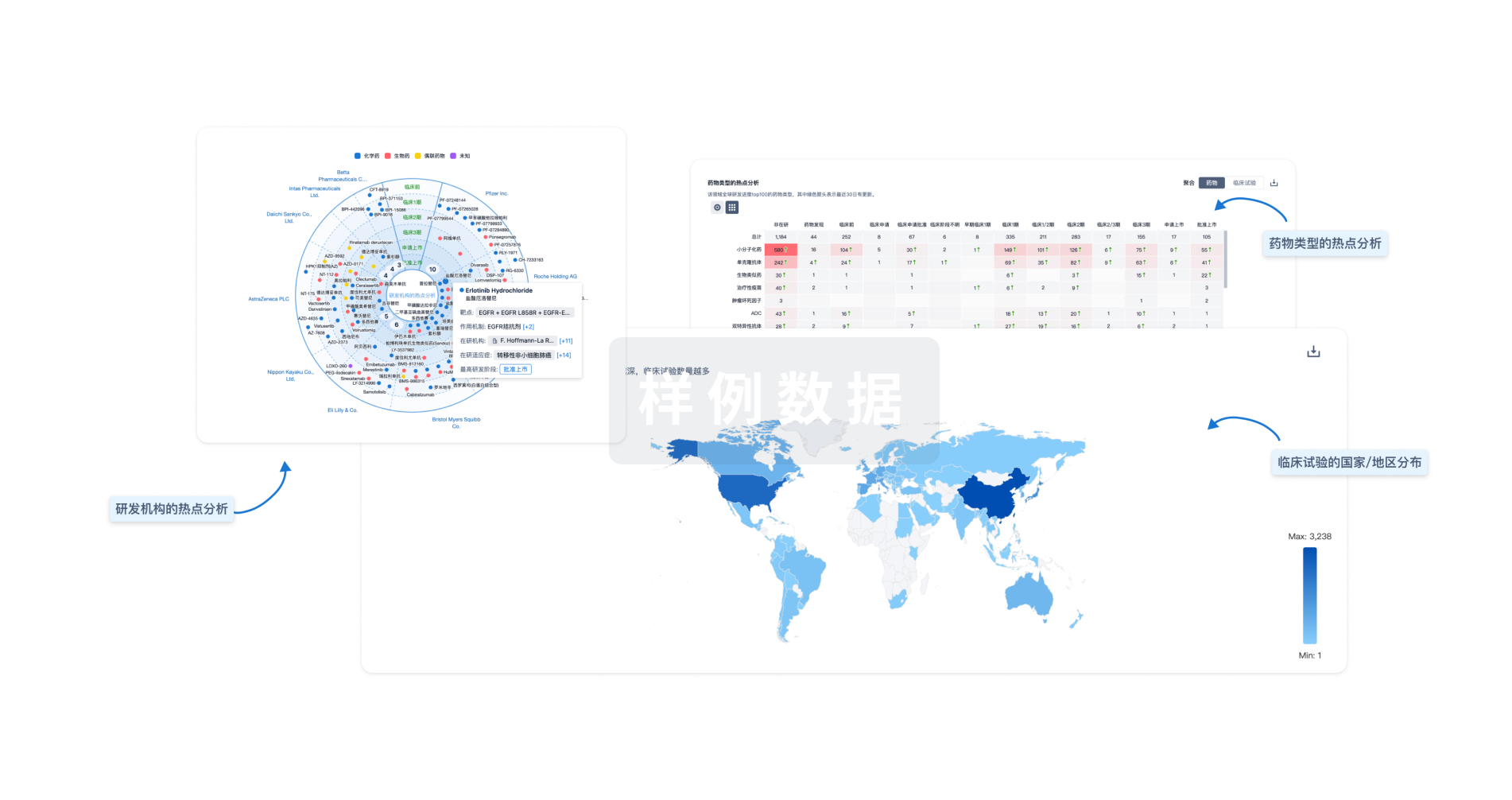
对领域进行一次全面的分析。
登录
或
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用
