预约演示
更新于:2025-05-07
Pterygium
翼状胬肉
更新于:2025-05-07
基本信息
别名 Corneal pterygium、Pterygium、Pterygium (disorder) + [30] |
简介 An abnormal triangular fold of membrane in the interpalpebral fissure, extending from the conjunctiva to the cornea, being immovably united to the cornea at its apex, firmly attached to the sclera throughout its middle portion, and merged with the conjunctiva at its base. (Dorland, 27th ed) |
关联
11
项与 翼状胬肉 相关的药物作用机制 CSF-1R拮抗剂 [+4] |
在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2014-10-15 |
靶点- |
作用机制- |
非在研适应症- |
最高研发阶段批准上市 |
首次获批国家/地区 日本 |
首次获批日期1987-10-01 |
靶点 |
作用机制 CaN抑制剂 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期1983-11-14 |
199
项与 翼状胬肉 相关的临床试验CTRI/2025/04/084537
Randomized controlled trial on Lodhradi Eyedrop Verses Moxifloxacin and Dexamethasone Eyedrop in post operative Patients of Pterygium (Prastari Arma) - NIL
开始日期2026-04-08 |
申办/合作机构- |
CTRI/2025/04/085563
An Open Randomized Control Clincal Trial To Evaluate The Efficacy Of Chaturbhadradi Varti Anjan In Prastari Arma With Special Reference To Progresssive Pterygium. - NIL
开始日期2025-05-01 |
申办/合作机构- |
CTRI/2025/04/084989
A Randomized Comparative Clinical Trial To Evaluate The Efficacy Of Elanir Kuzambu Anjana And Lauhadi Guggulu In The Management Of Arma (Pterygium) - NIL
开始日期2025-04-24 |
申办/合作机构- |
100 项与 翼状胬肉 相关的临床结果
登录后查看更多信息
100 项与 翼状胬肉 相关的转化医学
登录后查看更多信息
0 项与 翼状胬肉 相关的专利(医药)
登录后查看更多信息
3,578
项与 翼状胬肉 相关的文献(医药)2025-06-01·Experimental Eye Research
Genome-wide methylation analysis unveils genes and pathways with altered methylation profiles in pterygium
Article
作者: Prajna, N Venkatesh ; Prasad, Tejaswi ; Dharmalingam, K ; Devarajan, Bharanidharan ; Banerjee, Daipayan ; Aslam, Mohammed Hameed ; L, Mathan ; Gr, Aadhithiya T
2025-06-01·Cancer Epidemiology
A systematic review and meta-analysis of the association of human papilloma virus infections with ocular surface squamous neoplasia
Review
作者: Hall, Leanne ; Heal, Clare
2025-06-01·Experimental Eye Research
Construction of a competing endogenous RNA regulatory network in pterygium and role of hsa_circ_0081682 in fibroblast proliferation, migration, and apoptosis
Article
作者: Yang, Qiaodan ; Dou, Yulian ; Zhang, Ruiying ; Yan, Ming ; Tang, Xinyu
24
项与 翼状胬肉 相关的新闻(医药)2025-03-13
·抗体圈
核药领域正在进入收获期。
全球核药龙头捷报频传——诺华的两款核药2024年销售收入首次突破20亿美元;远大医药(0512.HK)交出的2024年答卷同样让人眼前一亮。
3月12日,远大医药发布2024年年度业绩,公司实现收入约116.45亿港元,剔除汇率同比增长12.8%;归母净利润约24.68亿港元,剔除汇率同比增长34%;得益于去年良好的业绩表现,公司拟派息每股0.26港元,合计超9.1亿港元。
这份成绩单发布后的首个交易日就获得了市场资金的强烈认可,远大医药股价一度大涨超过21%,最终当日收涨19.66%。
远大医药作为全球仅有的四家在肿瘤治疗方面成功实现创新核药商业化应用的创新制药企业之一,如今以易甘泰®钇[90Y]为首的核药板块已经成为公司收入增长最快的“尖兵”之一。同时,公司的全球化程度和核药管线数量在国际领先,目前拥有3款产品处在临床三期阶段,且核药产品储备数量已超过20个,未来像易甘泰®钇[90Y]这样的爆品,远大医药有望陆续推出。
立足当下,2025年将是远大医药业绩爆发的元年,公司凭借着“自研+合作引进+并购布局”多轮驱动投入转型创新的模式,2024年已经实现63项重大的里程碑,在年内新增14款商业化产品的同时,我们同时也看到公司的核药、五官科、呼吸等板块中的数款创新产品正在快步迈入商业化放量的新阶段,不仅将带动公司的收入规模快速增长,并且将公司整体的盈利结构和质量进一步优化提升。
远大医药即将迎来属于自己的股价和业绩的“戴维斯双击”时刻。
01
核药重磅炸弹时代来临
核药的重磅炸弹时代来临,远大医药将率先并充分受益。
2024年诺华两款核药收入达到21.16亿美元,其中Pluvicto实现销售13.92亿美元(同比增长42%),首次进入“10亿美元分子俱乐部”,而Lutathera得益于适应症范围的扩大(纳入了12岁及以上SSTR+胃肠胰神经内分泌肿瘤儿童患者),实现7.24亿美元(同比增长20%),在2-3年内也将进入10亿美元分子行列。
基于两款核药Pluvicto和Lutathera各自在前列腺癌、胃肠胰神经内分泌瘤领域展现出的强大治疗潜力和广阔的市场空间,近年来MNC和国际资本大力布局核药资产以“占位”下一个重磅炸弹,仅2024年MNC便完成了3笔核药并购交易,包括阿斯利康20亿美元收购Fusion Pharmaceuticals、BMS以41亿美元收购RayzeBio、诺华以10亿美元预付款+7.5亿美元里程碑收购Mariana Oncology。
远大医药在国内的强劲商业化表现,也正在兑现核药在全球市场所展现出的潜力。
2024年,远大医药核药板块实现收入约5.89亿港元,剔除汇率同比大幅增长近176.6%,其中易甘泰®钇[90Y]微球注射液自2022年国内获批上市后快速放量,已累计治疗近2000例患者,目前其正式手术已在国内22个省市的50余家医院展开。
易甘泰®钇[90Y]的快速放量,不仅基于远大医药出色的商业化,并且离不开核药自身治疗机制优势和优异的疗效解决临床未满足需求。
诺华的Pluvicto填补了PSMA阳性转移性去势抵抗性前列腺癌(mCRPC)治疗的临床空白,并在上市之初一度因产能问题导致供不应求。易甘泰®钇[90Y]在原发性肝癌和结肠癌肝转移领域为大量用药选择有限的患者提供了全新治疗选择,能够实现肝癌降期并帮助患者实现临床治愈,这是过去传统的治疗手段无法做到的。
易甘泰®钇[90Y]国内商业化以来已累计治疗近2000例患者,目前已有40余名患者顺利实现肝癌肿瘤降期转化并实施了肝癌切除手术,10余名患者通过桥接治疗成功实施肝移植,实现临床治愈。
易甘泰®钇[90Y]之外,随着远大医药与合作伙伴的快速临床推进,公司处于后期临床产品线正持续丰富,截至目前公司有3款核素偶联药物(RDC)处于临床三期,分别是前列腺癌诊断核药TLX591-CDx、透明细胞肾细胞癌诊断核药TLX250-CDx和胃肠胰神经内分泌瘤治疗性核药ITM-11,这也使得远大医药成为国内拥有最多三期临床创新核药管线的公司之一,足见公司在核药领域的深耕程度和布局的前瞻性。
这三款核药管线中TLX591-CDx、TLX250-CDx两款管线源自于与Telix的合作,而Telix在海外的表现已经充分证明了市场对其的认可和潜在巨大产品价值。
2024年,Telix总收入实现7.83亿澳元(约合4.87亿美元),同比增长56%;净利润约4990万澳元(约合0.31亿美元),同比增长860%。Telix业绩的高速增长的源于Illuccix®(即TLX591-CDx )的持续放量,自2022年Illuccix®获批以来,其在美国展现出强劲的放量表现。这种强劲的放量表现,不仅仅是得益于Pluvicto的快速爆量,更在于产品的高灵敏度和特异性,其不仅能够检测到90%以上原发性和转移性前列腺癌细胞表面的PSMA阳性病变,可用于检测基于血清PSA水平升高的复发前列腺癌患者,这促使Illuccix®在美国诊断性核药的竞争中更获得医生青睐。
自Illuccix®商业化放量以来,Telix的股价累计涨幅已经超过了500%。Telix管理层在最新的交流中也给出了2025年的业绩指引:公司收入将达到11.8~12.3亿澳元(同比增长50.6~57.05%),并计划继续增加研发投入,预计同比增长20~25%。
除此之外,Telix准备在2025年推出三款新产品 TLX007-CDx (Gozellix®)、TLX101-CDx(Pixclara®) 和 TLX250-CDx (Zircaix®),这对于远大医药来说显然是好消息,随着合作产品在美国实现商业化,国内管线可以通过桥接临床实现进度加速。
值得一提的是TLX250-CDx,目前透明细胞肾细胞癌(ccRCC)临床常用诊断方法CT或MRI提示存在肾脏肿块但无法判断是否为ccRCC的患者,TLX250-CDx正电子发射断层成像(PET)在诊断ccRCC的敏感性和特异性上分别达到86%和87%,远超过FDA要求的预设阈值,其有望成为ccRCC全新临床诊断标准。另外,TLX250-CDx并不局限于ccRCC,目前用于三阴性乳腺癌(TNBC)、非肌层浸润性膀胱癌(NMIBC)以及尿路上皮癌(UC)的多项适应症临床研究也在全球同步推进中。
在全球热门SSTR靶向核药布局方向,远大医药选择重点推进的治疗性核药ITM-11是一款潜在的同类最佳产品,这是一款将无载体177Lu与生长抑素类似物结合的RDC,全球三期临床COMPOSE研究显示,在侵袭性2/3级SSTR阳性的胃肠或胰腺来源的神经内分泌肿瘤GEP-NETs患者中,患者的mPFS高达34.5个月,且未观察到严重的不良事件。与Lutathera非头对头数据比较下,ITM-11有望在整体的疗效(mPFS显著提升,Lutathera一线治疗在22个月左右)、安全性上成为同类最佳产品。
远大医药不仅核药商业化拔得头筹,并且其核药诊疗一体化大平台已经逐渐成型,实现了自研&合作技术平台(20款储备管线)、温江研发生产基地、商业化平台的布局兑现,核药板块后劲很足。
02
新爆点:创新眼药攻势如潮
经历了数年的深耕与布局,2024年可谓是远大医药创新眼药的蓄力之年,未来几年眼科创新药物显然将成为公司的全新爆点。
过去,远大医药的五官科板块已经为公司在眼科商业化层面打下了坚实基础,已商业化的品种包括:国家保护独家品种的和血明目片(治疗眼底疾病)、治疗干眼症一线用药的人工泪液瑞珠等,这些均为畅销的大品种。
公司在多年的合作与沉淀后,已经构建起了一个丰富而具有差异化的创新眼药管线平台,从覆盖的疾病广度包括像屈光不正、干眼症这样的眼科大适应症,从差异化和未满足临床需求又可以覆盖到像蠕形螨睑缘炎、翼状胬肉这样的缺乏明确对症治疗药物的领域。
2024年至今,远大医药在创新眼药领域发生了许多重磅的里程碑事件,包括白内障术后抗炎镇痛创新药GPN00833完成国内三期临床研究并达到临床终点、从箕星药业引入OC-01和OC-02鼻喷雾剂两款干眼症药物的大中华区权益,以及蠕形螨睑缘炎创新眼药GPN01768的NDA获得CDE受理等,这几款创新药物未来的商业化放量都将给远大医药的眼药板块甚至公司整体业绩带来巨大的弹性。
博研咨询数据显示,2023年国内治疗药物市场高达120亿元,同比增长15%。其中,人工泪液在国内干眼症治疗市场中的占比超过50%。值得注意的是,当前市面上的人工泪液仅可模拟泪膜的一种或多种成分,难以真正对天然泪液进行替代。
远大医药已经商业化的OC-01(酒石酸伐尼克兰)是一种不含防腐剂鼻喷雾剂,其不同于人工泪液的眼表给药方式而采用创新的经鼻给药方式,其有效成份伐尼克兰通过结合烟碱型乙酰胆碱 (nACh) 使其活化,三叉神经副交感神经通路(控制泪膜自稳态)被激活,导致基础泪液分泌增加,从而达到治疗干眼的目的。
相比长期使用人工泪液可能产生的药物依赖、引发结膜炎和眼角膜损伤等安全性风险,OC-01拥有更高的选择性和安全性,其市场潜力非常可观。鉴于玻璃酸钠滴眼液、环孢素滴眼液在国内2023年销售规模均超过10亿元,OC-01凭借创新药属性和全新作用机制有望快速突破这一门槛。
远大医药前不久在三期临床取得成功的GPN00833是一款抗炎镇痛类激素纳米混悬滴眼液,目前激素眼用制剂是白内障手术后抗炎镇痛最常用且最有效的药物之一,可以快速有效控制眼科术后的感染、减轻患者眼部炎症和促进伤口愈合, 现有眼科术后围手术期激素眼用制剂(多为糖皮质激素,如地塞米松、泼尼松龙)普遍存在安全性风险较高的问题。
GPN00833利用独特的纳米制剂工艺解决了激素类产品低水溶性导致的生物利用度低及安全性风险,并在国内两项三期研究中其展现出可持久地清除眼部炎症和治愈眼部疼痛的疗效,同时安全性与安慰剂相似,有望对现有激素眼用制剂进行迭代。
博研咨询数据显示,2023年国内泼尼松龙滴眼液(专利期已过、单价35元/支)在8亿元左右。GPN00833作为安全性、有效性更优的迭代创新药,市场竞争格局更优、具备创新溢价,未来销售峰值超越泼尼松龙滴眼液只是时间问题。
处于NDA审评阶段的GPN01768(Xdemvy)是一种非竞争性γ-氨基丁酸氯离子通道(GABA-Cl)拮抗剂,用于治疗蠕形螨睑缘炎。蠕形螨睑缘炎是一种容易复发的疾病,影响的人群数量庞大,在中国和美国影响人群分别为4000万人和2500万人,在GPN01768获批之前全球尚未有针对蠕形螨睑缘炎的药物上市,临床空白巨大。
GPN01768在中国和美国的临床充分证明了自身对症杀螨的强大有效性和安全性,尤其国内三期临床研究LIBRA的顶线数据结果显示,GPN01768治疗蠕形螨睑缘炎患者的螨虫根除率具有统计学显著性(p<0.001),关键临床的大多数不良事件轻微可控,未出现治疗相关的停药。
GPN01768早在2023年7月在美国实现商业化,上市以来快速放量,2024年实现销售1.8亿美元,美银美林BofA更是预测GPN01768的峰值销售额将高达11亿美元。基于该药在美国上市以来的快速放量趋势、国内患者人群基数大于美国,其在国内的销售峰值潜力显然在10亿元之上。
集合上述分析,远大医药这三款商业化阶段创新眼药产品再加上治疗翼状胬肉的GPN00153(CBT-001)合计有着超越50亿国内销售的潜力,而这些差异化品种的合作引进,仅仅也只是公司前瞻性布局眼光和差异化品种战略的冰山一角的早期兑现,随着这些品种的商业化放量能力逐步验证以及公司自研产品的发力,远大医药的创新眼药板块成长性将会越来越强。
03
两大板块核心优势继续强化
远大医药布局特点是较快的并购节奏和明确指向补强,重点在于提升呼吸及危重症板块丰度,以及强化公司在心脑血管急救板块的竞争力。
呼吸、心脑血管这两大领域均为致死风险极高的疾病领域,临床未满足需求多,药物开发市场空间大。
2024年远大医药呼吸及危重症板块的表现突出,实现营收17.09亿港元,同比增长26.9%,展现了十足的成长力。这种稳定的成长力来源于公司深厚的积累,目前公司手握切诺®(黏液溶解促排剂)、恩卓润®和恩明润®(改善中重度哮喘)等稳定贡献现金流的品种,还有诸如Ryaltris®这样的创新品种正在蓄势待发。
在该领域,远大医药2024年完成了对百济制药的全资并购,在获得百济制药技术领先鼻喷制剂平台的同时,通过纳入该公司管线资产,形成了一套面向轻、中、重度过敏性鼻炎患者的吸入制剂药物产品组合。
远大医药治疗过敏性鼻炎鼻喷剂Ryaltris®正等待CDE的上市批准,作为国内第一个创新复方鼻喷剂其填补了一线对症治疗经治中重度过敏性鼻炎患者的临床空白。公司通过并购百济制药获得了一线过敏性鼻炎鼻喷剂用药(丙酸氟替卡松鼻喷雾剂、糠酸莫米松鼻喷雾剂、布地奈德鼻喷雾剂),这一产品组合未来要分享的是国内过敏性鼻炎百亿的用药市场,并购带来的化学反应不可估量。
另外,远大医药针对未满足临床需求如重症治疗监护、呼吸窘迫综合征、脓毒症等领域开发了拥有自主知识产权的“全球新”药物,如脓毒症创新药STC3141已完成国内二期临床患者入组并给药、脓毒症创新药APAD已完成了国内一期临床并顺利达成临床终点。
在心脑血管急救板块,远大医药近年来品种数量正在快速扩充(心脑血管急救板块从2022H1的24个品种到2024年的30个品种),产品管线数量位居行业前列,覆盖血压控制、血管活性药、心肌代谢、心衰、抗凝等领域,后劲充足。
2024年,远大医药该板块力美通®依普利酮片成功进入国家医保目录,进一步促进了产品可及性的提升。同时,公司通过布局天津田边合心爽®、合贝爽®这样的畅销心脑血管急救药物(临床上治疗冠状动脉痉挛心绞痛的首选药物)迅速切入如如糖尿病、胃肠道疾病等慢性疾病赛道,同时获得天津天边的制剂产能加速公司原料制剂一体化,提升公司的盈利能力。另外,远大医药同年收购了多普泰,其核心产品脉血康系列药物(销售规模在5亿元)及其他产品与公司现有产品组合也形成了良好的协同效应。
随着远大医药的创新急救产品Jext®在大湾区获批上市,叠加这两大并购资产的注入,未来几年将成为公司全新增长动力,带来一揽子有充分竞争力的产品,为板块业绩成长提供坚实保障。
从对远大医药的呼吸及危重症、心脑血管急救这两大优势板块布局分析不难看出,公司在过去的投入及积累了大量的现金牛产品,已经逐渐放量成为各板块业绩的稳定内生增长动力;而公司通过自研、引入具备差异化的创新产品,同时辅以补强指向明确的资产并购,使得公司在呼吸、心脑血管这两大板块未来不缺乏“强大的发动机”,这显然能够双轮驱动公司板块以及整体业绩快速提升。
结语:历经多年的投入、前瞻性的布局眼光和强大商业化并购整合能力,远大医药的核药、眼药、呼吸和心脑血管等几大板块正在快速步入创新布局兑现期。核药板块易甘泰®钇[90Y]有望进一步放量冲击10亿单品,成都温江甲级核素研发生产基地将在2025年建成形成集研发、生产、运营为一体的全产业链版图;呼吸及危重症板块,STC3141国内脓毒症二期有望在上半年取得初步成药性数据;五官科板块,新一代干眼症药物商业化加速放量,多款创新眼药将迎来重大里程碑;另外,心脑血管精准介入、生物科技板块,公司也在丰富产品技术布局和积极拓展商业化链路,为公司增长贡献稳健增量。
可以预见的是,远大医药2024年交出的这一份出色的业绩答卷也仅仅是这艘巨轮加速行驶的预兆,而更多的惊喜和爆发式成长或许还在后头,我们或许能见证一家传统Pharma转型创新成功的涅槃过程。
识别微信二维码,添加抗体圈小编,符合条件者即可加入抗体圈微信群!
请注明:姓名+研究方向!
本公众号所有转载文章系出于传递更多信息之目的,且明确注明来源和作者,不希望被转载的媒体或个人可与我们联系(cbplib@163.com),我们将立即进行删除处理。所有文章仅代表作者观点,不代表本站立场。
并购临床3期财报申请上市
2025-02-11
Walter and Eliza Hall Institute of Medical Research in Victoria, Australia led one of the largest eye studies in the world that used AI to analyse eye images of over 50,000 people to better understand the retina's connection with various diseases.
The international study, funded by California-based Lowy Medical Research Institute, tapped into a dataset of optical coherence tomography (OCT) images from around 54,000 individuals stored at the UK Biobank. Utilising a convolutional neural network, it generated "the highest-resolution spatial dataset of retinal thickness ever produced," creating 50,000 maps with measurements at over 29,000 locations across the retina.
Moorfields Eye Hospital and University College London from the United Kingdom and the University of Washington in the United States were collaborators in the study.
FINDINGS
While previous studies have indicated correlations between retinal thickness and diseases, the WEHI-led research provided a deeper look into the complex anatomy of the retina.
One of the significant findings of this study, published in Nature Communications, is that reduced retinal thickness is "highly associated" with multiple sclerosis (MS), a chronic neurological disease affecting the brain and spine. "This result provides strong, independent confirmation of multiple reports of the utility of OCT as the source of biomarkers for MS and MS progression," it added.
Retinal thinning is also highly associated with a range of other neurodegenerative diseases, as well as cardio-metabolic disorders.
"We illustrated that the retina has unique metabolic sensitivities, with retinal thickness associated with multiple systemic metabolic diseases, and metabolites previously implicated in several retinal diseases," the study explained.
Additionally, the study identified genetic factors that influence retinal thickness, suggesting that at least 294 genes play a role in the retina's growth and development.
WHY IT MATTERS
Researchers claim their study opens up the potential for using routine eye imaging to screen for and manage diseases. Neurodegenerative conditions like dementia and metabolic disorders such as diabetes are linked to degeneration or disruption of the central nervous system, which the retina is a part of.
"We’ve shown that retinal imaging can act as a window to the brain, by detecting associations with neurological disorders like multiple sclerosis and many other conditions," lead researcher Dr Vicki Jackson from WEHI.
The study specifically points to the potential for retinal thickness as a diagnostic biomarker "to aid in detecting and tracking the progression of numerous diseases."
"We can now pinpoint specific locations of the retina which show key changes in some diseases," Dr Jackson said.
Moreover, the study contributes to the growing field of oculomics as a "non-invasive approach for predicting and diagnosing diseases."
THE LARGER TREND
Another Australian study has linked the damage to the conjunctiva of the eye to pterygium, or tissue growth on the cornea, which can be an early predictor of skin cancer. A desktop and mobile AI-driven detection system has been developed to assess sun-related eye damage.
The Singapore Eye Research Institute has also utilised AI to scan people's retinal photos and assess their health conditions. Over the past five years, it developed two novel solutions: one for screening chronic kidney disease and another for predicting a person's biological age.
Meanwhile, in China, a generative AI model was built for automated eye disease diagnosis. Called VisionFM, it was pre-trained using 3.4 million eye images from over 500,000 people worldwide. Besides diagnosing eye diseases, it can be applied to eye disease progression prediction, systemic biomarker prediction through ocular imaging, intracranial tumour prediction, and lesion, vessel, and layer segmentation.
2024-12-18
·药智网
国内创新含量最高的眼药企业之一开始展露锋芒。
远大医药作为国内眼科创新药管线布局最多的公司之一,近日创新眼药资产布局和临床进展频频,预示着公司在眼药领域的创新布局加速进入成果兑现期。
图片来源:瞪羚社
11月4日,远大医药白内障术后抗炎镇痛创新药GPN00833完成国内三期临床研究并达到临床终点,预示着其将成为继公司蠕形螨睑缘炎创新眼药GPN01768(TP-03)之后第二个完成国内三期并快步进入商业化的创新眼药产品。
12月11日,远大医药与箕星药业达成合作,公司获得治疗干眼症的全球首创创新产品酒石酸伐尼克兰鼻喷雾剂“OC-01”和“OC-02”(Simpinicline)鼻喷雾剂在大中华区(中国大陆及中国港澳台地区)独家开发及商业化的权益,公司在干眼症的布局再下一城。值得注意的是,OC-01是目前全球首款且唯一一款获批治疗轻、中、重度干眼的无防腐剂、多剂量、无菌包装鼻喷雾剂。
其后,远大医药向CDE递交的蠕形螨睑缘炎创新眼药GPN01768新药上市申请(NDA)获得了受理,GPN01768作为公司首个从国内临床阶段布局并推进到上市审评创新眼药产品,这一消息预示着公司创新眼药布局即将进入商业化阶段。
远大医药如此密集的创新眼药里程碑催化及公司在创新资产布局上的重拳出击,不仅为公司眼科创新药板块商业化加速及落地提供了强力支持,同时让本就拥有人工泪液“瑞珠”和血明目片等多个畅销在售眼药品种的远大医药“创新含量”和盈利能力有了指数级的提升。
国内创新眼药龙头,名副其实。
01
干眼商业化布局再下一城
远大医药布局箕星药业干眼症产品组合,无疑增加了公司在干眼症领域产品梯队厚度和市场核心竞争力。
目前,远大医药在干眼症领域拥有人工泪液产品“瑞珠”(聚乙烯醇滴眼液,主打无防腐剂策略),在国内人工泪液品种中销售名列前茅。
此次交易属于对现有主流用药品种“迭代更新”。
目前干眼症临床上最常用的治疗方案为人工泪液,市面上常见的人工泪眼产品多为眼表给药方式的滴眼液产品,然而天然泪液成分较为复杂(超过1500种成分),市面上人工泪液仅可模拟泪膜的一种或多种成分,难以真正对天然泪液进行替代。
箕星药业的OC-01(酒石酸伐尼克兰)是一种不含防腐剂鼻喷雾剂,其不同于人工泪液的眼表给药方式而采用创新的经鼻给药方式,其有效成份伐尼克兰通过结合烟碱型乙酰胆碱(nACh)受体而发挥作用。当受体活化后,三叉神经副交感神经通路(控制泪膜自稳态)被激活,导致基础泪液分泌增加,从而达到治疗干眼的目的。
OC-01三期临床结果显示,相比对照组,OC-01在改善干眼症患者泪液分泌方面显示出具有统计学和临床意义的显著改善,受试者的自然泪液分泌较基线明显增加(Schirmer评分较基线增加大于或等于10毫米的受试者比例显著占优)。显然,OC-01市场上传统干眼症治疗方法不同,这款产品拥有非常高的选择性和安全性,很大程度上降低了长期用药带来的副作用。
干眼症作为眼科药物的“兵家必争之地”,其蕴含的市场潜力惊人。
在所有的眼科疾病中,干眼症是发病率排名第二(国内发病率21%-30%),仅次于屈光不正,国内医疗机构门诊就诊的干眼症患者占眼科总就诊人数的30%以上。以目前主流用药人工泪液玻璃酸钠滴眼液和环孢素滴眼液为例,2023年销售规模均突破10亿元,呈现出迅速放量的趋势。今年8月,欧康维视生物大手笔从爱尔康引进8款干眼症治疗和手术用滴眼液产品组合大中华权益,昭示着行业玩家对干眼症领域的高度重视。
OC-01早在2021年10月就被FDA批准在美国上市,其长期的患者安全性数据显然为国内后续商业化提供了强有力的参照和推广证据。另外,OC-01也在2024年11月被CDE批准上市,远大医药作为在干眼症领域深耕多年的商业化眼药龙头,获得OC-01权益之后能够迅速进展商业化工作,进一步提升眼药商业线销售人效,同时增强了公司在干眼症产品线厚度。值得注意的是,另一款布局的干眼症产品OC-02也刚完成了在海外IIb期干眼症临床,和OC-01一样同属经鼻给药的高选择性乙酰胆碱能受体激动剂。
02
两大创新单品,
快速冲刺商业化
GPN01768、GPN00833将是远大医药前两个率先进入商业化的创新眼药产品,分别瞄准蠕形螨睑缘炎、白内障术后抗炎镇痛两大适应症市场,呈现出补充未满足临床需求和对现有治疗药物降维打击的潜力。
睑缘炎是一种常见的眼科疾病,其特征是眼睑边缘发炎、发红和眼部刺激等临床表现。蠕形螨眼睑炎是由蠕形螨感染引起的,约占所有睑缘炎病例的2/3以上,以眼痒、眼异物感、眼干、睑缘充血等为典型临床表现。这种疾病对睑缘的破坏较大,易反复发作,导致脂质分泌减少,泪膜稳定性下降,进而引发干眼。更严重时会导致角膜炎症,给患者造成严重的视力损伤。
过去,全球缺乏安全有效的蠕形螨睑缘炎治疗药物,主要治疗方式多样,包括睑缘清洁、眼部热敷、睑板腺按摩及强脉冲光(IPL)治疗等,但大多数杀螨效果有限且缺乏有力的临床证据支持。
远大医药的GPN01768是一款新型滴眼液(0.25%洛替拉纳),即一种非竞争性γ-氨基丁酸氯离子通道(GABA-Cl)拮抗剂,其通过选择性抑制蠕形螨体内的GABA-Cl,使虫体麻痹和死亡,进而治疗蠕形螨睑缘炎。
GPN01768在中国和美国的临床充分证明了自身对症杀螨的强大有效性和安全性,在海外两项Saturn-1、Saturn-2研究中(合计833例蠕形螨性睑缘炎),前者蠕形螨根除比例红疹治愈比例分别为68%和19%,远优于安慰剂组的17%和7%。后者给药组蠕形螨根除比例红疹治愈比例分别为50%和30%,安慰剂组只有14%和9%;中国蠕形螨睑缘炎患者的三期临床研究LIBRA的顶线数据结果显示,GPN01768治疗蠕形螨睑缘炎患者的螨虫根除率具有统计学显著性(p<0.001)。更重要的是,上述关键临床的大多数不良事件轻微可控,未出现治疗相关的停药情况。
GPN01768(TP-03)于2023年7月获得美国FDA批准上市,是FDA批准的唯一一款针对蠕形螨睑缘炎的药物;GPN01768在2023年取得了1470万美元的销售收入,在2024年前三季度,GPN01768在美国的销售量已达到约12万瓶,总收入约达1.3亿美元,放量迅速。美银美林BofA更是预测GPN01768的峰值销售额将高达11亿美元。
GPN01768之所以拥有如此广阔的市场空间,基于蠕形螨性睑缘炎较大的患者基数和疾病易复发需反复用药的特性。数据显示,蠕形螨性睑缘炎可能影响多达2500万美国人,每年至少有4500万人去眼科诊所就诊;尽管GPN01768能有效地去除眼睛上的螨虫,但螨虫会在身体其他部位生存,并最终迁移回眼睛,临床研究中大约40%的患者在12个月后会复发,需重新用药。
中国目前有超过4000万蠕形螨睑缘炎患者,比美国患者人群数量更大,并且国内尚未有针对蠕形螨睑缘炎的药物上市,GPN01768上市后有望填补该临床空白。另外,蠕形螨也是睑板腺功能障碍的危险因素之一,目前中国有超过7000万睑板腺功能障碍患者。GPN01768在美国开展的用于治疗蠕形螨导致的睑板腺功能障碍的二期临床研究也显示出了阳性的顶线结果。
远大医药作为GPN01768大中华区权益的拥有者,参照GPN01768在美国快速商业化放量的成功及国内当前大量未满足的临床需求,GPN01768很可能成为远大医药创新眼药管线中冲击10亿元的重磅品种之一。
另一方面,近期国内临床三期成功的GPN00833是一款抗炎镇痛类激素纳米混悬滴眼液,其活性成分丙酸氯倍他索一种强效的糖皮质激素,GPN00833利用独特的纳米制剂工艺解决了激素类产品低水溶性导致的生物利用度低及安全性风险,剑指现有眼科术后围手术期的抗炎镇痛药物迭替代。
激素眼用制剂是目前白内障手术后抗炎镇痛最常用且最有效的药物之一,可以快速有效控制眼科术后的感染、减轻患者眼部炎症和促进伤口愈合。
然而,受眼用制剂技术能力限制,目前国内激素眼用制剂由进口产品主导,该细分市场近十年无新产品上市。今年三月,GPN00833获得FDA批准在美国上市,其为15年来全球首个获批的激素类纳米制剂。
以现有已上市的激素眼用制剂如地塞米松、泼尼松龙等来看,其成分多为传统糖皮质激素,此类药物长期应用不良反应较多,可能导致眼压升高,引发激素性青光眼、激素性白内障等并发症,从而诱发或加重感染,市场中急需疗效&安全性高的强效激素滴眼液。
GPN00833有望填补当前迫切存在的临床需求,在海外两项三期研究中,GPN00833耐受性良好且安全性与安慰剂相似。有效性层面,国内三期研究结果显示:GPN00833可迅速且持久地清除眼部炎症和治愈眼部疼痛,效果在临床及统计学上均显著优于安慰剂。经GPN00833治疗,31.8%的受试者在术后第8天至第15天的前房细胞计数为0,而安慰剂组为20.0%。此外,91.2%的GPN00833组的受试者在术后第4天即达到无眼痛状态并维持至第15天无复发,显著高于安慰剂组的55.3%,且安全性良好。
可以预见,GPN00833在国内的商业化前景一片光明。博研咨询数据显示,2023年国内泼尼松龙滴眼液在8亿元左右。由于该药物专利期已过(单价35元一支),GPN00833作为安全性、有效性更优的迭代创新药,市场竞争格局更优、具备创新溢价,未来销售峰值大概率能超越泼尼松龙滴眼液。另外据东北证券预测,未来几年国内白内障手术量将呈现每年10+%的稳步增长,眼科术后镇痛药物市场规模有望加速扩大。
图片来源:东北证券
远大医药GPN01768、GPN00833这两款创新眼药,通过对现有未满足临床需求的差异化、迭代布局,在商业化落地后有望收获两个10亿级别的现金牛品种。
03
远大眼药的布局雄心
过去,市场只听闻远大医药的眼药品种很多、销售规模很大,但显然没有意识到公司是“大而强”和“不仅强而创新属性拉满”。
随着GPN01768、GPN00833加速向商业化阶段进发和公司将已商业化的OC-01权益收入囊中,可以预见远大医药在两年内将在国内拥有三款上市的创新眼药产品,并且这三款产品均拥有冲击10亿级别销售的能力。
放眼国内,拥有三款创新眼药爆品的,远大医药很可能是第一家上市公司。同时,随着创新药板块布局的开花结果,公司该板块的盈利能力和价值将产生质变影响;更重要的是,远大医药原本就具备眼科产品线和商业化推广团队,眼科创新药的加入,无论从销售的人效来说还是眼科创新药本身的利润率和产品的竞争格局看,都能给远大眼科板块带来极大的正向反馈。
远大医药在眼药的布局,还不止于上述的创新管线。
干眼症作为国内发病率第二高的眼病,远大医药在该领域竞争力已足够强。公司作为国内眼药龙头,雄心显然不满足于此,将布局延伸至第一大发病率眼病“近视防控”。
远大医药通过产学研转化布局了一款用于治疗近视防治的新型眼用制剂GPN00884,与现有普遍使用的低浓度阿托品相比,GPN00884滴眼液不会出现瞳孔散大效应,不会出现畏光、调节下降等不良反应,给药时段不受限制,可提高患者的依从性,在2024年3月国内已进入临床阶段。
除此之外,和GPN01768一样远大医药聚焦未满足临床需求差异化布局了治疗翼状胬肉创新药物CBT-001。目前全球还未有对症治疗翼状胬肉的药物获批,而翼状胬肉在中国和美国影响的患者分别超过1亿人和1500万人,CBT-001是目前全球临床进度最快的药物(预计在2025年底前完成全球多中心临床,包括中国和美国),其他竞争药物均在临床早期。同时,CBT-001已有临床二期数据表明,其能够高效的减少翼状胬肉血管及结膜充血、抑制角膜新生血管病变,安全性良好。有海外机构预测CBT-001销售峰值在6-8亿美元,考虑到国内定价略低于海外但患者群体庞大,该药物未来市场潜力同样巨大。
分析远大医药眼科创新药管线不难看出,公司该领域的创新布局非常有章法,一方面立足于眼科的大适应症进行纵深布局,另一方面通过追求在未满足的眼病临床需求上布局“First in class”、布局更优异疗效的创新药“迭代或降维打击”现有传统用药来实现差异化商业化竞争,以换取更广阔的市场空间和极优的竞争格局,这也不禁让投资者赞叹公司布局的前瞻性和眼光。
不仅如此,公司创新管线的布局也得到了稳固的销售能力的支持。作为远大医药重点布局的战略发展方向之一,公司在眼科领域已吸引和培养了一批兼具临床经验和营销经验的专业人士,建立了专业化营销团队,并与大型医药流通企业和连锁药店建立了长期稳定合作,形成了覆盖全国的营销网络。随着研发的推进,未来三年公司有望实现多款创新产品的上市,为其持续健康发展提供新动能。
从另外一个角度看,远大医药创新转型的布局不仅仅局限在眼药领域。近期以来,远大医药在核药、危重症等领域的临床进展捷报频传。
以核药领域为例,远大医药用于诊断肾透明细胞癌的创新RDC药物TLX250-CDx国内三期临床研究完成首例患者入组给药、用于治疗胃肠胰腺神经内分泌瘤的创新RDC药物ITM-11国内三期临床研究申请或药监局受理。通过长期的深耕布局,远大医药目前已成为拥有进入中国三期临床研究中诊断和治疗类RDC创新药总计储备最多的企业,也是全球范围内在核药抗肿瘤领域拥有最丰富产品管线和诊疗一体化布局的创新药企之一。同时,公司也在积极推进国内甲级资质核素生产平台的建设,目前已完成了主体结构封顶,未来有望进一步加速其全球创新研发管线的落地。
远大医药在其多个业务领域都陆续实现了像前述的眼药板块呈现出产品群阶段性开花结果的态势,比如公司创新核药易甘泰上市以来呈现爆发式放量、过敏性鼻炎多个新产品未来将集体发力助力板块规模快速放大等,这均为公司在创新药和创新转型超前、持续投入布局带来的正向结果。
结语:创新眼药素来是拥有超强爆发力的产品,比如海外罗氏双抗Vabysmo在2022年初才推出2023年销售额就超过24亿瑞士法郎、兴齐眼药低浓度阿托品在2024年上市后首个季度销售便有1.7亿元。远大医药作为国内拥有创新眼药产品最多的龙头,其未来创新眼药国内销售规模有望开启爆发式的增长,进一步巩固公司创新眼药龙头的地位。
创新眼药板块的开花结果,仅仅也只是远大医药坚定投入布局创新药物水到渠成的结果,而公司未来的业绩增长点不止于眼药板块,而是在核药、呼吸及危重症板块、创新器械等多个板块上演。
友情推荐:医药行业深度技术内容,点击“博药”查看详情~
来源 | 瞪羚社(药智网获取授权转载)
撰稿 | Kris.
责任编辑 | 八角
声明:本文系药智网转载内容,图片、文字版权归原作者所有,转载目的在于传递更多信息,并不代表本平台观点。如涉及作品内容、版权和其它问题,请在本平台留言,我们将在第一时间删除。
合作、投稿 | 马老师 18323856316(同微信)
阅读原文,是受欢迎的文章哦
临床3期医药出海申请上市临床成功
分析
对领域进行一次全面的分析。
登录
或
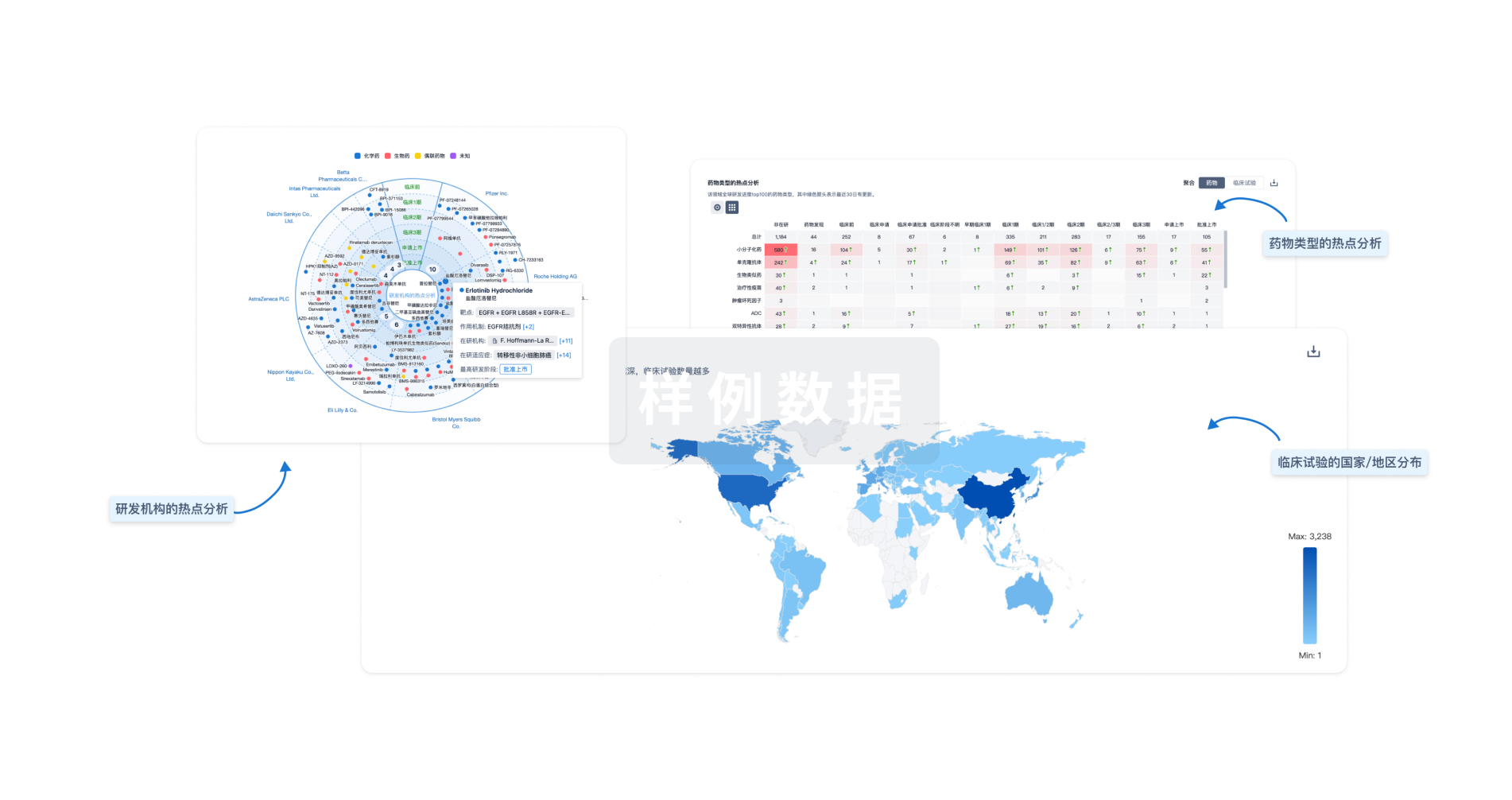
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用

