预约演示
更新于:2025-05-07
Localized obesity
局部肥胖
更新于:2025-05-07
基本信息
别名 Fatty tissue hyperplasia、Hyperplasia, fatty tissue、Lipohyperplasia + [18] |
简介- |
关联
10
项与 局部肥胖 相关的药物靶点 |
作用机制 GPCR激动剂 |
在研机构 |
在研适应症 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期2015-04-29 |
作用机制 β2-adrenergic receptor激动剂 |
原研机构 |
最高研发阶段批准上市 |
首次获批国家/地区 美国 |
首次获批日期1994-02-04 |
64
项与 局部肥胖 相关的临床试验CTRI/2025/04/084882
A Single Arm Clinical study to assess the efficacy ofPaschimottanasana in the management of Central Obesity - NIL
开始日期2025-05-26 |
申办/合作机构- |
CTRI/2025/03/082120
Add on effect of khadira Asana bhavitha thriphala choorna with selected yoga techniques in central obesity among post menopausal women - NIL
开始日期2025-03-27 |
申办/合作机构- |
NCT06756490
A Prospective, Blinded (Evaluators and Subjects Blinded), Randomized Controlled Phase I Clinical Trial to Evaluate the Safety, Efficacy and Pharmacokinetics of Liposome-encapsulated Deoxycholic Acid for Injection (HY-2003) Administered by Subcutaneous Fat Layer Injection in the Subjects With Excessive Submental Fat Accumulation
The main purpose of this study is to evaluate the safety of HY-2003 in subjects with moderate to severe submental fat accumulation at different doses and dosing frequencies in comparison with the positive control and placebo. The secondary objectives include: Evaluating the pharmacokinetic characteristics of HY-2003 after a single administration in humans and obtaining preliminary pharmacokinetic parameters; Evaluating the efficacy of HY-2003 under different doses and dosing frequencies, compared with the positive control and placebo, in the treatment of subjects with moderate to severe submental fat accumulation.
开始日期2024-12-26 |
申办/合作机构  四川汇宇制药股份有限公司 四川汇宇制药股份有限公司 [+2] |
100 项与 局部肥胖 相关的临床结果
登录后查看更多信息
100 项与 局部肥胖 相关的转化医学
登录后查看更多信息
0 项与 局部肥胖 相关的专利(医药)
登录后查看更多信息
330
项与 局部肥胖 相关的文献(医药)2025-04-21·Zhonghua wai ke za zhi [Chinese journal of surgery]
[Application of magnetic recanalization technology in treatment of complex refractory benign biliary stricture].
Article
作者: Liu, X M ; Lyu, Y ; Xu, H ; Zhang, X F ; Li, Y
2025-04-01·Gene
Alström syndrome: A rare cause of dilated cardiomyopathy in five Chinese children
Article
作者: Sun, Ningning ; Wang, Ziwei ; Yin, Jie ; Qi, Yuying ; Wang, Yuqi ; Lu, Jie ; Zhou, Jueru ; Wang, Chunli ; Yang, Shiwei
2025-03-01·Trends in Molecular Medicine
Bone–brain crosstalk in osteoarthritis: pathophysiology and interventions
Review
作者: Tang, Yilan ; Wang, Zhiyan ; Tu, Yiheng ; Cao, Jin
2
项与 局部肥胖 相关的新闻(医药)2025-01-23
The FDA adjusted the risk of the medicine after identifying 82 cases worldwide from 1996 to 2024. Image credit: Shutterstock / Jeppe Gustafsson.
The US Food and Drug Administration (FDA) is adding the risk of anaphylaxis to a new boxed warning of glatiramer acetate, a medicine for multiple sclerosis (MS).
The agency issued the warning about the rare but serious risk of allergic reaction to the medicine, known under the brand names Copaxone and Glatopa, via a drug safety communication on 22 January. The products already have warnings of immediate post-injection reaction, chest pain, localised fat loss, and immune response modification, as per the drugs’ labels.
Teva Pharmaceutical’s Copaxone was approved by the FDA in 1996 to treat relapsing forms of MS, an autoimmune disorder that affects the central nervous system. Glatopa, a generic version of Teva’s product, was approved by the FDA this year and marketed by
Sandoz
. The drugs have demonstrated a 34% drop in the number of relapses compared to placebo.
The FDA adjusted the risk of the medicine after identifying 82 cases worldwide across the last 28 years. The agency said that although rare compared to how often the medicine is used, the serious allergic reactions meant emergency room visits or hospitalisations were required in all patients in the dataset. A majority of the 82 patients experienced anaphylaxis within one hour of taking the medicine and six died.
The FDA’s alert follows a similar notification from the UK’s Medicines and Healthcare products Regulatory Agency (MHRA) in October 2024, which outlined the risk of anaphylactic reactions. The UK regulator said that allergy-related events could occur shortly after administration or take months or years after treatment initiation.
A multiple sclerosis patient from Castlebar in Ireland died shortly after administering herself with Copaxone in June 2024, according to local newspaper
Mayo Live
. A post-mortem concluded she died of anaphylactic shock and a ‘suspected adverse reaction report was submitted to Ireland’s Health Products Regulatory Authority (HPRA).
Identifying anaphylaxis associated with glatiramer acetate can be challenging because its initial symptoms are similar to immediate post-injection reaction, a temporary and common side effect. The FDA recommended healthcare professionals to educate patients on the signs and symptoms of post-injection reactions and told patients to immediately stop taking glatiramer acetate in the event of breathing difficulties, face or throat swelling, or rash appearance.
Neither Teva nor Sandoz has publicly commented about the new boxed warning.
Teva faced difficulties with the European Commission with Copaxone in November 2024. The agency
fined Teva €462.6m ($481m)
after concluding that the company artificially extended the therapy’s patent protection and disseminated misleading information about rival products.
上市批准专利到期专利侵权
2022-05-30
GAITHERSBURG, Md. and SUZHOU, China, May 30, 2022 /PRNewswire/ -- Sirnaomics Ltd. (the "Company" or "Sirnaomics", stock code: 2257.HK), a leading biopharmaceutical company in discovery and development of RNAi therapeutics, announced today the launch of a Phase I clinical trial of the Company's siRNA (small interfering RNA) drug candidate, STP705, in adults undergoing abdominoplasty for submental fat reduction. This study is the first application for Sirnaomics to apply an RNAi therapeutic candidate for medical cosmetology treatment.
The global non-invasive fat reduction market size was valued at USD1.1 billion in 2021 and is anticipated to register a compound annual growth rate (CAGR) of 16.1% from 2022 to 2030, up to approximately USD4.0 billion (Grand View Research). Non-invasive fat reduction is a procedure that is done to decrease or eliminate stubborn fat pockets in specific areas of the body by using methods like cryolipolysis, radio frequency, and laser lipolysis. Non-invasive devices that are used in these procedures to permanently abolish fat cells are approved by the U.S. Food and Drug Administration (FDA) as their efficiency and safety have been tested and the results have been proven to show significant results. However, the market is currently dominated by devices that have mild therapeutic effects and require multiple treatment sessions. Sirnaomics will initially focus on the use of STP705 for submental fat reduction.
The dose-ranging, randomized, double-blind, vehicle-controlled study will enroll 10 patients to evaluate the safety and tolerability of STP705, which will be delivered via subcutaneous injection. The primary endpoints are to assess injection comfort, characterize local and systemic safety, and evaluate histological changes of subcutaneous doses of STP705, and to compare the safety and tolerability of three different concentrations of STP705 to select dosages for future studies.
"Submental fullness is a common condition that is resistant to diet and exercise and is influenced by multiple factors including aging and genetics," said Sirnaomics Executive Director and Chief Medical Officer Michael Molyneaux M.D. "Our goal in expanding our applications of STP705 with this study is to examine the safety and efficacy of this treatment in patients who are undergoing abdominoplasty. We hope to use the information from this study to expand into the treatment of submental fat reduction and other areas of non-invasive fat sculpting. This Phase I study will serve as a blueprint for future studies of STP705 in the medical aesthetics category."
"The current devices and procedures for non-invasive fat reduction have mild therapeutic effects, and require multiple and sometimes challenging treatment sessions, which are costly for patients and don't always achieve the desired results," said Dr. Patrick Lu, Founder, Chairman of the Board, Executive Director, President and CEO of Sirnaomics. "Based on a discovery from our previous clinical study and a series of preclinical evaluations with animal models, we decided to expand STP705 therapeutic application into medical aesthetics with this Phase I study."
About STP705
Sirnaomics' leading product candidate, STP705, is a siRNA (small interfering RNA) therapeutic that takes advantage of a dual-targeted inhibitory property and polypeptide nanoparticle (PNP)-enhanced delivery to directly knock down both TGF-β1 and COX2 gene expression. The product candidate has received multiple IND approvals from both the U.S. Food and Drug Administration (FDA) and the Chinese National Medical Products Administration (NMPA), including treatments of cholangiocarcinoma, non-melanoma skin cancer and hypertrophic scar. STP705 has also received Orphan Drug Designation for treatment of cholangiocarcinoma (CCA) and primary sclerosing cholangitis (PSC). STP705 is currently in five clinical trials for different indications: a Phase IIa for squamous cell carcinoma in situ (isSCC), a Phase II for basal cell carcinoma (BCC), a Phase I/II for keloid scarring, a Phase I/II for hypertrophic scar (HTS), and a Phase I for liver cancer (basket).
About Sirnaomics
Sirnaomics is an RNA therapeutics biopharmaceutical company with product candidates in preclinical and clinical stages that focuses on the discovery and development of innovative drugs for indications with medical needs and large market opportunities. Sirnaomics is the first clinical-stage RNA therapeutics company to have a strong presence in both China and the United States, and also the first company to achieve positive Phase IIa clinical outcomes in oncology for an RNAi therapeutics for its core product, STP705. Learn more at www.sirnaomics.com.
临床结果孤儿药创新药siRNA
分析
对领域进行一次全面的分析。
登录
或
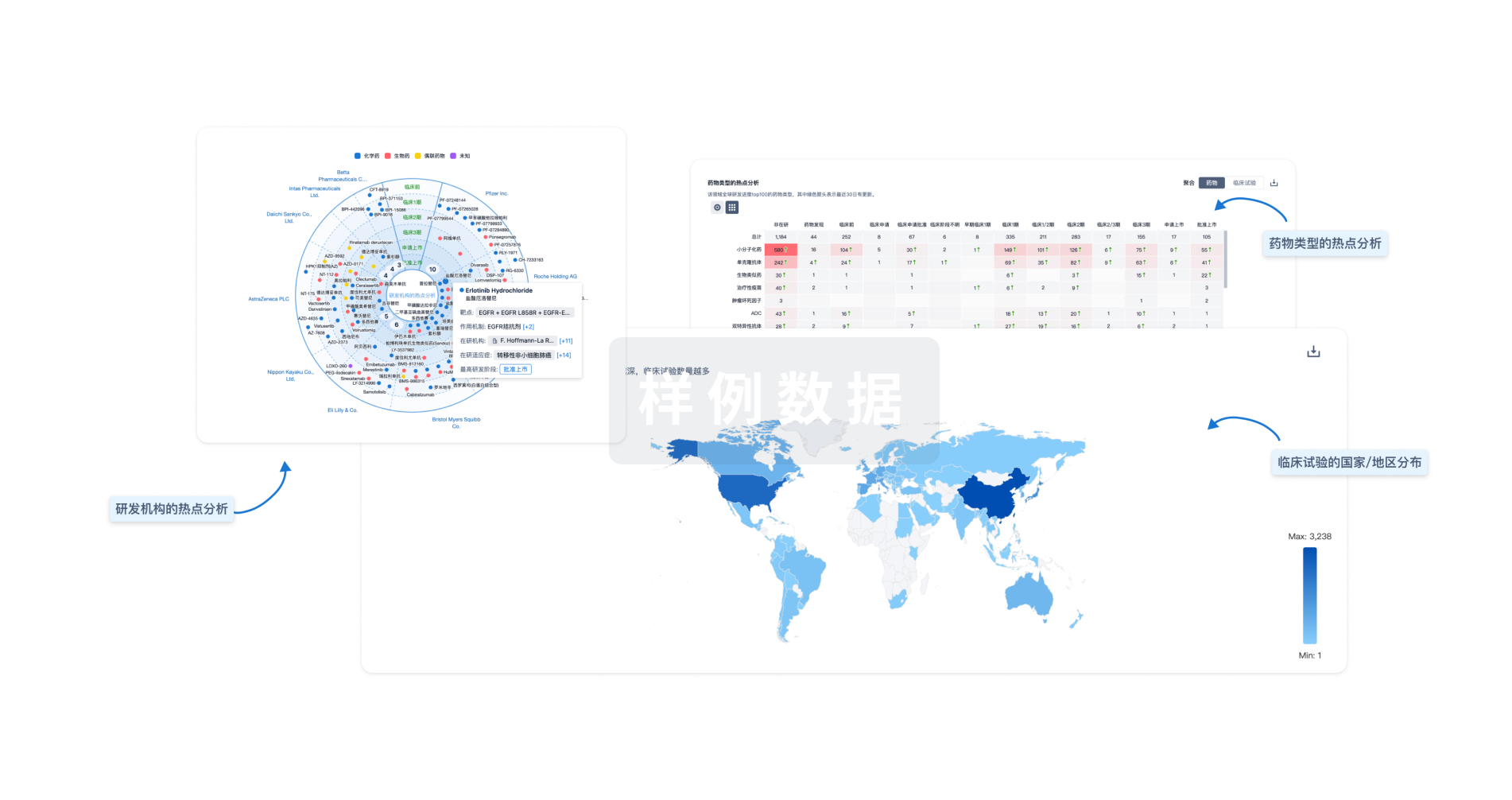
生物医药百科问答
全新生物医药AI Agent 覆盖科研全链路,让突破性发现快人一步
立即开始免费试用!
智慧芽新药情报库是智慧芽专为生命科学人士构建的基于AI的创新药情报平台,助您全方位提升您的研发与决策效率。
立即开始数据试用!
智慧芽新药库数据也通过智慧芽数据服务平台,以API或者数据包形式对外开放,助您更加充分利用智慧芽新药情报信息。
生物序列数据库
生物药研发创新
免费使用
化学结构数据库
小分子化药研发创新
免费使用


